Re:Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis
작성자문형철작성시간20.01.21조회수2,099 목록 댓글 0beyond reason
Review Article | Open Access
Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis
Abstract
Reactive oxygen species- (ROS-) induced lipid peroxidation plays a critical role in cell death including apoptosis, autophagy, and ferroptosis. This fundamental and conserved mechanism is based on an excess of ROS which attacks biomembranes, propagates lipid peroxidation chain reactions, and subsequently induces different types of cell death. A highly evolved sophisticated antioxidant system exists that acts to protect the cells from oxidative damage. In this review, we discussed how ROS propagate lipid peroxidation chain reactions and how the products of lipid peroxidation initiate apoptosis and autophagy in current models. We also discussed the mechanism of lipid peroxidation during ferroptosis, and we summarized lipid peroxidation in pathological conditions of critical illness. We aim to bring a more global and integrative sight to know how different ROS-induced lipid peroxidation occurs among apoptosis, autophagy, and ferroptosis.
1. Introduction
Reactive oxygen species (ROS) are produced by normal physiological processes and play important roles in cell signaling and tissue homeostasis [1]. However, excess radical species produce adverse modifications to cell components and augment various pathogenesis, such as lipids, proteins, and DNA damage [2]. Cellular membranes or organelle membrane, due to their high polyunsaturated fatty acids (PUFAs), are especially susceptible to ROS damage, which is called “lipid peroxidation.” Lipid peroxidation is a process in which free radical species such as oxyl radicals, peroxyl radicals, and hydroxyl radicals remove electrons from lipids and subsequently produce reactive intermediates that can undergo further reactions. The lipid peroxidation damages phospholipids directly and can also act as cell death signal which induces programmed cell death. Oxidized phospholipids can also play an important role in many inflammatory disease and frequently mediate proinflammatory change [3]. Recently, ferroptosis, a new form of programmed cell death, has been found to be caused by lipid peroxidation [4], which highlights lipid peroxidation during the physiological process of cell death. It is therefore of great interest to understand how ROS is produced and eliminated and how ROS-induced lipid peroxidation contributes to cell death. In this review, we summarize the processes of ROS-induced lipid peroxidation among apoptosis, autophagy, and ferroptosis and discuss how they come together to affect the fate of a cell in a more global and integrative way.
2. Generation of ROS and Antioxidant System
2.1. Generation of ROS
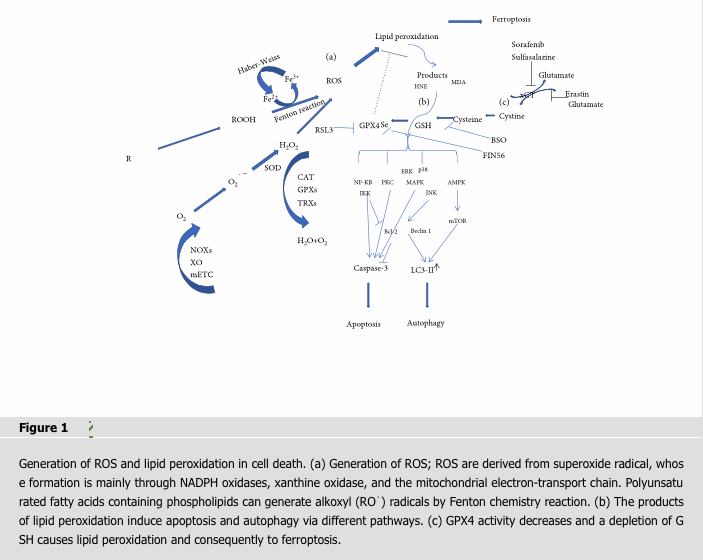
ROS are partially reduced oxygen-containing molecules, which are free radicals and/or oxygen derivatives, including superoxide anion, hydrogen peroxide, hydroxyl radical, lipid hydroperoxides, and peroxyl radicals. Most intracellular ROS are derived from superoxide radical, whose formation is mainly through NADPH oxidases (NOXs), xanthine oxidase (XO), and the mitochondrial electron-transport chain (mETC) in endogenous biologic systems [5, 6]. ROS are converted to hydrogen peroxide by the superoxide dismutase (SOD) and yield the highly toxic hydroxyl radical in the presence of reduced iron (Fe2+) through the Fenton reaction which have different peroxide species to generate hydroxyl (⋅OH) or alkoxyl (RO⋅) radicals [7]. Ferric iron (Fe3+) can be recycled to Fe2+ via the Haber-Weiss reaction by oxidation with a peroxyl radical to oxygen [8, 9] (Figure 1). Imbalance in the rate of ROS generation leads to oxidative stress and consequent production of free radicals that can damage DNA, proteins, and lipids [10].
2.2. Antioxidant System
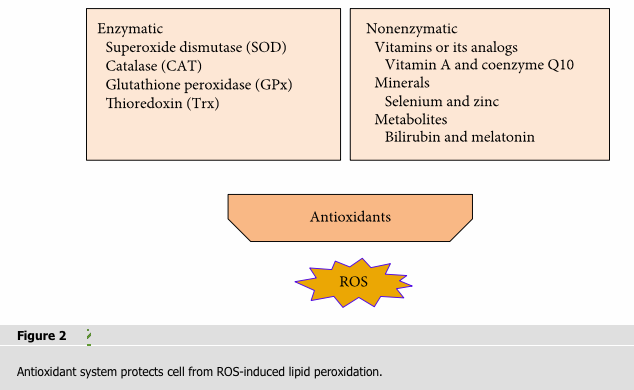
Antioxidants can counteract free radicals and neutralize oxidants. The general endogenous antioxidant system consist of (1) enzymatic antioxidants like superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx), and thioredoxin (Trx); and (2) nonenzymatic antioxidants that include vitamins or its analogs (vitamins A, C, and E; coenzyme Q10; and flavonoids), minerals (selenium and zinc), and metabolites (bilirubin and melatonin) (Figure 2).
2.2.1. Enzymatic Antioxidants
The enzymatic antioxidants have an effective protective effect against oxidative attack due to the ability to decompose ROS [11]. Among them, SOD is very important for living cell because most of ROS is produced from superoxide. SOD can catalyze the conversion of superoxide into oxygen and hydrogen peroxide [12]. CAT can decompose the hydrogen peroxide into molecular oxygen and water. CAT significantly reduced oxidative stress and restored mitochondrial structure by enhancing the mitochondrial membrane potential (Δψm) so as to play an antiapoptotic effect and normalize replicative and wound healing capacity [13, 14]. Glutathione peroxidases (GPxs) consist of multiple isoenzymes with distinct subcellular locations exhibiting different tissue-specific expression patterns [15, 16]. During persistent oxidative stress, GPxs become the main H2O2-scavenging enzymes after ascorbate peroxidases being inhibited [17]. GPxs detoxify other toxic organic hydroperoxides by catalyzing the reduction of H2O2 and hydroperoxides to water or alcohols [16]. The thioredoxin (Trx) antioxidant system is composed of NADPH, thioredoxin reductase (TrxR), and Trx. Trx and TrxR catalyze the NADPH-dependent reduction of the active-site disulfide in oxidized Trx to give a dithiol in reduced Trx. NADPH maintains CAT in the active form and is used as a cofactor by TRX and GSH reductase, which converts glutathione disulfide (GSSG) to glutathione (GSH), a cosubstrate for the GSH-Pxs. The reaction sequence of TrxR-mediated reduction of protein disulfides has been reviewed [18]. The thioredoxin (Trx) and glutathione (GSH) systems could be complementary to each other. The TrxR1/Trx1 system can sustain reduced GSH pools in the absence of glutathione reductase [19].
2.2.2. Nonenzymatic Antioxidants
Vitamin A (retinol) is a carotenoid synthesized in the liver and resulted from the breakdown of β-carotene. Vitamin A can directly interact with peroxyl radicals forming free carbon-centered radical adducts and scavenge peroxyl radicals by electron transfer before they propagate peroxidation to lipids [20]. Coenzyme Q10 (CoQ10) is the single lipophilic antioxidant which is essential for electron transport during mitochondrial respiration. It was reported to prevent oxidative damage of lipid peroxyl radicals and improve mitochondrial biogenesis [21, 22]. Vitamin C (ascorbic acid) can scavenge a variety of oxygen free radicals [23]. Vitamin E (a-tocopherol) is a fat-soluble antioxidant that can protect the polyunsaturated fatty acids (PUFAs) in the membrane from oxidation, regulate the production of ROS, and modulate signal transduction [24]. Flavonoids are extensively distributed in beverages, vegetables, and fruits which have antioxidant activity by inhibiting the enzymes responsible for superoxide production as well as NADH oxidase [25]. Selenium and zinc have the antioxidative function due to their activity maintenance of many enzymes. Zinc acts in the stabilization of membranes by inhibiting NADPH-oxidase and inducing the synthesis of metallothioneins, and it is also a component of SOD [26]. Selenium serves as a structural and catalytic cofactor for numerous proteins such as GPxs and thioredoxin reductase (TrxR) which is an important component of enzymatic antioxidants. Many metabolites such as bilirubin and melatonin have an antioxidative function. Evidence suggests that bilirubin possesses antioxidant properties. Bilirubin treatment can inhibit the TLR4-mediated upregulation of iNOS by preventing activation of hypoxia inducible factor-1α (HIF-1α) through scavenging of NOX-derived ROS [27]. Melatonin improves the intramitochondrial antioxidative defense by enhancing reduced glutathione levels and inducing glutathione peroxidase and superoxide dismutase to inhibit peroxidation [28]. Melatonin may prevent mitochondrial damage for it behaves like synthetic mitochondrion-targeted antioxidants which concentrate in mitochondria at relatively high levels [29].
3. Production of Lipid Peroxidation and Its Detecting Methodologies
ROS generation in the biomembranes is very high due to the solubility of molecular oxygen. Thus, the membrane phospholipids, containing high levels of PUFAs, are extremely sensitive to be attacked by ROS [30]. Moreover, the PUFAs themselves convert into reactive free radicals after reacting with the free radicals which are able to propagate lipid peroxidation chain reactions [31].
3.1. Production of Lipid Peroxidation
The products of lipid peroxidation chain reactions display high biological activity [32]. It destroys DNA, proteins, and enzyme activity as well as acts as molecular to activate signaling pathways initiating cell death [33]. Biomembranes are prone to undergo lipid peroxidation, and it is possibly via two pathways: nonenzymatic and enzymatic.
3.1.1. Nonenzymatic Autoxidation
The nonenzymatic pathway which is also called “nonenzymatic phospholipid (PL) autoxidation” is iron-dependent lipid peroxidation. Autoxidation radical chain reactions of PUFA containing PLs can be divided into three stages: initiating—polyunsaturated acyl chain of a PL is oxidized to generate R⋅ (a carbon-centered radical containing PL) by losing hydrogen to hydroxyl (⋅OH); propagating—R⋅ readily reacts with molecular oxygen to form a peroxyl radical (R-OO⋅) [34]. On the one hand, propagation reactions of R-OO⋅ include hydrogen abstracting from a PL molecule to form a lipid hydroperoxide (R-OOH). On the other hand, addition of R-OO⋅ to the bis-allylic position of another PL forms R-OO-R⋅ dimers [35]. During the process of Fenton chemistry, R-OOH can undergo reductive cleavage to generate alkoxyl (RO⋅) radicals [36]. Autoxidation reactions of PUFAs form many electrophilic species such as malondialdehyde, isoprostanes, and 4-hydroxy-2-nonenal (4-hydroxy-2,3-trans-nonenal, HNE). These products of lipid peroxidation have various biological functions [37]. The last stage is terminating—two radicals of the chain reaction react with each other to form stable molecules and the antioxidants effectively decompose radicals which inhibit the chain reaction [38] (Figure 3).
3.1.2. Enzymatic PL Peroxidation
Enzymatic peroxidation is catalyzed by lipoxygenase (LOX) which can also participate in the formation of R-OOH. Lipid peroxidation involves a highly organized oxygenation center, wherein oxidation occurs on only one class of phospholipids [39]. Arachidonic (C20: 4) and linoleic (C18: 2) are the most abundant polyenoic fatty acids that serve as substrates for LOX using molecular oxygen to form hydroperoxyl groups at different carbon position of acyl chains [40]. Arachidonate lipoxygenase-15 (Alox15) which encodes for the 12/15-LOX has a unique substrate requirement among the LOX family. It can directly oxygenate PUFA containing PLs without prior release of esterified PUFA by phospholipase A2 (PLA2) [41]. Alox5 and Alox12 which encodes for a 5-lipoxygenating and enzyme platelet-type 12-LOX, respectively, have been shown to provide oxygenated acyl precursors to generate oxygenated PLs [42, 43].
3.2. Detecting Methodologies
Various methods have been applied to the measurement of oxygen radicals and their damaging effects on membrane lipids. Hydroperoxides, primary product of lipid peroxidation, are not stable which may include phospholipid hydroperoxides [44]. An HPLC-chemiluminescence (HPLC-CL) detection method has been developed by which these species can be measured [45]. The main aldehyde product of lipid peroxidation is the 3-carbon dialdehyde species malondialdehyde (MDA) and 4-hydroxy-2-nonenal (HNE) [46]. MDA can be measured by the thiobarbituric acid (TBA) test using UV/visible spectrophotometry. Immunoblotting or immunohistochemistry is available for measuring 4-HNE/protein adducts [47]. Isoprostanes (IsoP) are a series of prostaglandin-like compounds produced by a free radical-mediated lipid peroxidation of arachidonic acid independent of cyclooxygenase. Utilizing gas chromatography/mass spectrometry (GC/MS) with negative ion chemical ionization, the lipid peroxidation-derived 5- and 15-F2t isoprostanes (8-isoprostaglandin F 2α) can be accurately measured in biological fluids which has been regarded as the most reliable approach for accessing lipid peroxidation in vivo [48]. An alternate immunoassay (ELISA) for 8-isoprostaglandin F 2α in biological tissues is provided [49]. In recent years, the use of fluorescent probes specifically designed to detect oxidative stress in living cells, probes based on dihydrofluorescein diacetate, has been applied in vitro. A lipophilic fluorescent dye 4,4-difluoro-5-(4-phenyl-1,3-butadienyl)-4-bora-3a,4a-diaza-s-indacene-3-undecanoic acid (C11-BODIPY 581/591) to probe oxyl-radical-induced lipid oxidation by flow cytometry (FCM) has higher sensitivity and specificity for lipid peroxidation in vitro [50].
4. Roles of Lipid Peroxidation in Different Cell Death
4.1. Lipid Peroxidation in Apoptosis
Apoptosis is programmed series of events dependent on energy, as well as morphological features such as cell shrinkage, chromatin condensation, and presence of apoptotic bodies without inflammatory reactions [51, 52]. There are mainly three alternative pathways that lead to apoptosis: (1) extrinsic pathway, (2) intrinsic pathway, and (3) perforin/granzyme pathway. Caspases are key molecules involved in the transduction of the apoptosis signal, and all of the pathways converge to the executioner caspase-3 [53]. The extrinsic pathway is initiated by the tumor necrosis factor (TNF) receptor family interacting with a ligand and then binds with procaspase-8 following ligand-receptor interaction to activation of caspase-3 which leads to execution of apoptosis [54, 55]. The intrinsic pathway (mitochondrial pathway) employs alterations of inner mitochondrial membrane for induction of apoptosis. Apoptosis is triggered when the Bcl2-family proapoptotic proteins cause the opening of mitochondrial permeability transition pore and proapoptotic proteins into cytoplasm by interacting with apoptotic protease-activating factor 1 (Apaf-1) and procaspase-9 to constitute apoptosome [56, 57]. An assembly of apoptosome leads to caspase-9 activation, which further activates caspase-3, for apoptotic execution [58]. Perforin/granzyme-induced apoptosis is employed specifically by CD8+ cytotoxic T cells used by cytotoxic lymphocytes to eliminate virus-infected or transformed cells [59]. Granzyme B in the vesicles could activate procaspase-10 or directly activates caspase-3 for execution of apoptosis [60]. Granzyme A cleaves an inhibitory complex of a DNAse to induce apoptosis [61].
Lipid peroxidation play an important role in apoptosis. The products of lipid peroxidation interacts with membrane receptors and transcription factors/repressors to induce signaling for apoptosis. It can stimulate the activation of both the intrinsic and extrinsic apoptotic signaling pathways [62, 63]. ROS may lead to cardiolipin peroxidation, a mitochondrion-specific inner membrane phospholipid, and subsequent products of lipid peroxidation formation activated intrinsic apoptosis [64]. We next summarized different signal pathways of activation of apoptosis by the products of lipid peroxidation.
4.1.1. The Products of Lipid Peroxidation Induce Apoptosis via Different Signal Pathways
The NF-κB protein family is widely involve in inflammation, stress response, survival, and cell death [65]. Previous studies demonstrated that the product of lipid peroxidation increased NF-κB activity by inhibiting IκB degradation [66]. Besides the action of the product of lipid peroxidation on IKK, one of the NF-κB pathway element was shown to phosphorylate antiapoptotic Bcl-2 for its inactivation upon lipid peroxidation [67]. It is also known that NF-κB are also responsible for the transcriptional regulation of antiapoptotic expression [68]. Based on this background, lipid peroxidation was proposed to regulate the NF-κB pathway, the antiapoptotic Bcl-2, and the crosstalk between these survival elements.
Mitogen-activated protein kinases (MAPKs) are also responsible for cellular signal transduction in response to a diverse set of stimulators including oxidative stress [69]. The activation of MAPKs causes the phosphorylation of serine, threonine, and tyrosine residues of proteins to executive regulating function. The extracellular signal-regulated kinase (ERK), p38, and Jun N-terminal kinase (JNK) can activate MAPKs under various conditions which affects cytoprotective or apoptotic signaling [70]. It has been shown that the product of lipid peroxidation forms adducts with ERK, JNK, and p38 to activate MAPKs for the activation of caspase signal initiating the apoptotic processes [71–73].
The protein kinase C (PKC) is a key regulator of a plethora of the transduction of cellular signals that regulate cell proliferation, differentiation, and apoptosis [74]. PKC isoforms are activated by growth factors by the stimulating phospholipase C (PLC), which generates inositol trisphosphate (IP3) and diacylglycerol (DAG) [75]. Many PKC isoforms are lipid-sensitive and Ca2+-dependent enzymes. The product of lipid peroxidation stimulates PKC indirectly through the activation of phospholipase C or affecting the activity of its subunits [76]. It has been suggested that the product of lipid peroxidation can activate protein kinase C-delta (PKCδ), a member of the lipid-regulated serine/threonine PKC family, preventing triglyceride accumulation in obese mice [77]. PKCδ is cleaved by caspase-3 to generate a constitutively activated catalytic fragment, which amplifies apoptosis cascades [78]. Thus, lipid peroxidation can activate the PKC pathway to regulate apoptosis.
4.2. Lipid Peroxidation in Autophagy
Autophagy, the process of cellular self-eating, is an important protein degradation pathway, especially during stress conditions. It is known as a cellular catabolic pathway that plays crucial roles in cellular homeostasis including the maintenance of cellular function and viability [79]. There are three main autophagic pathways: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) [80, 81]. Chaperone-mediated autophagy involves the direct translocation of cytosolic proteins across the lysosomal membrane while microautophagy involves inward invagination of lysosomal membrane delivering a small portion of cytoplasm into the lysosomal lumen [82]. Among the three types of autophagy, the most extensively studied is macroautophagy which is mediated by a special organelle termed the autophagosome. During the process, light chain 3 (LC3) is involve in the formation of autophagosomes in mammalian cells that serves as a biomarker for occurrence of autophagy [83]. A selective form of macroautophagy in which mitochondria are specifically targeted for degradation at the autophagolysosome is so called mitophagy [84]. Mitophagy plays an essential role in cell differentiation, programming, cell death, and immune response [85]. Mitochondrial damage and dysregulation of mitophagy have been implicated in neurodegenerative diseases, cancer, and cardiac disease [86–88]. Autophagy helps to remove the damaged mitochondria and oxidized proteins, in most cases, supports survival. Under normal conditions, ROS-induced autophagy reduces damage caused by oxidative stress to protect cells. For instance, autophagy plays a protective role by eliminating ROS so as to preserve the integrity of mitochondria, prevent apoptosis, and promote antigen presentation. However, excessive autophagy induced by ROS can also cause autophagic cell death under certain circumstances [89].
A complex and differential regulation of autophagy by lipid peroxidation has been suggested by several studies [90–92]. The products of lipid peroxidation can adduct to specific mitochondrial and autophagy-related proteins driving cellular dysfunction in an autophagic cell death way [90]. During myocardial ischemia and reperfusion, autophagy signaling such as AMP-activated protein kinase and Akt-mTOR signaling is compromised by the products of lipid peroxidation through interference with upstream regulators [91]. Lipid peroxidation products may induce lysosomal dysfunction and lipofuscinogenesis which results in reduced autophagy activity [92]. We will summarize different signal pathways of activation of autophagy by the products of lipid peroxidation.
4.2.1. The Products of Lipid Peroxidation Trigger Autophagic Cell Death via Different Signal Pathways (Figure 1)
The AMPK/mTORC pathway initiates autophagy. Adenosine monophosphate-activated protein kinase (AMPK) as upstream regulators of the mammalian target of rapamycin (mTOR) pathway senses nutrient and energy depletion and activates the tuberous sclerosis complex (TSC1–TSC2), leading to mTOR inactivation and initiation of autophagy [93]. The mammalian TOR (mTOR) pathway which negatively regulates macroautophagy is present in all types of lysosomes [94]. Inhibiting mTORC1 signaling by rapamycin, a major sensitive inhibitor of mTORC, significantly increased the level of LC3-II and led to autophagy [95]. It is suggested that the production of lipid peroxidation may activate mTORC1 signaling through direct inhibition of AMPK. The production of lipid peroxidation could conjugate with liver kinase B1 (LKB1), an upstream substrate of AMPK, and thus result in the activation of the mTOR pathway in isolated cardiomyocytes [96].
The JNK-Bcl-2/Beclin 1 pathway initiates autophagy. C-Jun N-terminal protein kinase (JNK), a member of the MAPK family, mediates Bcl-2 phosphorylation and Bcl-2 dissociation from Bcl-2/Beclin 1 complex which functions in the lysosomal degradation pathway of autophagy [97, 98]. JNK activation-induced phosphorylation of Bcl-2 plays an important role in Bcl-2-dependent autophagy without inactivation of the mTOR pathway which constitutes a distinct molecular signature of autophagy [99]. The production of lipid peroxidation could promote its interaction with JNK as a result of the nuclear translocation of this kinase to stimulate autophagy [100].
4.3. Lipid Peroxidation in Ferroptosis
Ferroptosis is a new form of programmed cell death characterized by iron-dependent increase in ROS [101]. Ferroptosis plays crucial roles in cellular proliferation, senescence, and differentiation. A study has shown that senescent cells were highly resistant to ferroptosis due to iron accumulation [102]. A specific ferroptosis inhibitor, ferrostatin-1 (Fer-1), has been used to evaluate the role of ferroptosis in various pathophysiological settings. Fer-1 could prevent oxidative lipid damage and could delay cyst development in polycystic kidney disease, suggesting the necessity of ferroptosis for cell proliferation in fibrosis-related disease [103]. Deletion of the Gpx4, one of the ferroptosis-executing gene, neuron-like cell became more sensitive to ferroptosis upon differentiation. These results reinforce the susceptibility of neuronal context to ferroptosis and suggest the value of ferroptosis in neuroprotection [103].
Ferroptosis can be triggered by structurally diverse small molecules (e.g., erastin, sulfasalazine, and RSL3) and also prevented by lipophilic antioxidants (CoQ10, Vitamin E, ferrostatins, and liproxstatins) [4, 8, 39, 104]. Ferroptosis occurs as a result of increased ROS levels due to elevated intracellular iron concentration and a depletion of antioxidant GSH that cause lipid peroxidation and consequently to cell death [101]. More and more studies have confirmed that GPX4 activity decreases or iron excess leads to ferroptosis [105–107]. Ferroptosis is distinct from apoptosis, autophagy, and other modes of cell death. How lipid peroxidation leads to ferroptosis is still an unsolved mystery [108]. Thus, in this review, we only focus on the mechanism of lipid peroxidation during ferroptosis related to both GPX4 activity and iron metabolism.
4.3.1. GPX4 Activity Affecting Lipid Peroxidation Leads to Ferroptosis
GPX4 is an antioxidant enzyme that neutralizes lipid peroxides and protects membrane fluidity by using glutathione, as a cofactor of GPX4, to protect cells and membranes against peroxidation. Oxidized glutathione disulfide (GSSG) is subsequently reduced by glutathione reductase and NADPH/H+ to recirculate reduced glutathione (GSH) [109]. Inhibiting GPX4 can lead to increased ROS [110], while overexpression of GPX4 can reduce ROS and subsequently prevent cell from ferroptosis [111, 112]. GPX4 is a specific and robust central regulator of ferroptotic cell death when it is directly inhibited or indirectly inactivated by depletion of glutathione [112] (Figure 1).
4.3.2. GPX4 Inhibitors and Selenium Influence GPX4 Activity Directly
RSL3 is a GPX4-specific inhibitor. Analysis of mass spectrometry-based proteomic data from an affinity pull-down experiment ranked GPX4 (PHGPx) as the top protein target for RSL3 [112, 113]. Inhibiting GPX4 by RSL3 generates lipid ROS and induces ferroptosis [114]. Selenium-containing GPX4 is important for living cell to allow the utilization of biomembranes for increased cellular plasticity and for the utilization of peroxides as second messengers in redox signaling processes [115, 116]. Without selenium, GPX4 lost its activity and cells are highly sensitive to oxidative damage due to irreversible overoxidation of the catalytically active-site thiolate [117]. FIN56 could decrease GPX4 abundance and derived production of coenzyme Q10. Studies have shown that FIN56 is not a cystine/glutamate antiporter inhibitor because it does not affect GSH levels. Instead, FIN56 treatment resulted in loss of GPX4 protein through posttranslational degradation and blocked mevalonate-derived production of lipophilic antioxidants such as coenzyme Q10 [118] (Figure 1).
4.3.3. GSH Influence GPX4 Activity Indirectly
GPX4 plays an antioxidant effect by catalyzing its substrate—GSH. Thus, GSH influence GPX4 activity indirectly. Glutathione biosynthesis catalyzed by GCL (glutamate-cysteine ligase) and GS (glutathione synthetase) is essential for maintaining redox homoeostasis which needs cysteine, glutamate, and glycine as substrates [119]. Thus, the intracellular concentration of these substrates and the activity of enzymes affect GSH production.
Inhibiting cystine-glutamate antiporter decreases the intracellular concentration of cysteine which affects levels of GSH. The cystine/glutamate antiporter solute carrier family 7 member 11 (SLC7A11; also known as xCT) is a component of a plasma membrane transporter which is responsible for extracellular cystine and intracellular glutamate exchanging [120]. Erastin abolished the import of cysteine which is a precursor for glutathione during ferroptosis, and it was also proved to be a potent, selective inhibitor of system xc- [4, 121]. Sorafenib, a multikinase inhibitor, is an FDA-approved drug used for treating advanced hepatocellular carcinoma [122]. Compared to other kinase inhibitors, sorafenib is the only drug that displays ferroptotic efficacy [123]. Similar to erastin, sulfasalazine had also been repurposed to induce ferroptotic cancer cell death via increased accumulation of lipid ROS [124].
Glutamate decreases the intracellular concentration of cysteine mediating ferroptosis. Glutamate induces oxidative stress via the inhibition of cysteine transporter xCT, leading to depletion of the cellular glutathione pool [125]. Inhibiting glutamate-induced toxicity can be initiated by calcium influx after glutamate receptor activation [126] or by competitive inhibition of a systemxc-dependent process, suggesting that ferroptosis is involved [127] (Figure 1).
Buthionine sulfoximine (BSO) induced ferroptosis by suppressing glutathione levels [113]. BSO is an inhibitor of glutamate-cysteine ligase, the rate limit in genzyme for glutathione synthesis [128]. Glutathione depletion causes loss of cellular antioxidant capacity and inhibition of glutathione-dependent enzymes such as glutamate-cysteine ligase. BSO was demonstrated to suppress glutathione levels and induce ferroptosis by inhibiting GCL [128] (Figure 1).
4.3.4. The Production of ROS during Iron Metabolism Causes Lipid Peroxidation
Iron is required for numerous critical processes such as DNA synthesis, heme synthesis, and iron-sulfur cluster synthesis [129, 130]. It also plays an important role in the active sites of various enzymes which are involved in the formation such as LOX, xanthine oxidase, NADPH oxidases, and mitochondrial complex I and III [131–133]. However, the levels of iron in the cell need to be tightly balanced, as an excess of iron can impair cellular functions due to the generation of ROS and eventually cell death [106]. The increases of ROS caused lipid peroxidation and ferroptosis, which was suppressed by the treatment with iron chelator deferoxamine [8]. In addition, a higher level of iron transport proteins increased iron-mediated ROS and subsequently led to ferroptosis [134, 135]. Iron-mediated ROS was important to ferroptosis, and the production of ROS during iron metabolism was mainly from the process of “Fenton reaction” and Haber-Weiss reaction [8, 9]. Additionally, cells contain small amounts of uncoordinated and redox-active Fe2+, the so-called “labile iron pool” (LIP) [136]. Lysosomes can recycle endogenous iron sources like ferritin and mitochondria, and it is also a particularly large LIP [137]. Thus, lysosomes play a great important role in iron metabolism. Inhibitors of iron metabolism and iron chelators (e.g., deferoxamine (DFO) and ciclopirox (CPX)) suppress lipid peroxidation by reducing the availability of iron from iron pool [8]. Accordingly, both Fenton chemistry and iron-dependent enzymes may generate the reactive forms of oxygen that can trigger lipid peroxidation and finally cause ferroptotic cell death (Figure 1).
5. Lipid Peroxidation in Critical Illness
Critical illness including acute kidney injury, severe sepsis, and cardiac injury is interwoven with inflammation and oxidative stress, and the consequent production of lipid peroxidation plays an important role in the progression of disease. Abundant experimental and clinical data supported the important role of lipid peroxidation-related mechanisms in critical illness by reducing or genetic mutation to modulate lipid peroxidation. Inhibiting lipid peroxidation by Fer-1 can prevent folic acid- (FA-) induced acute kidney injury in mice which associates with downregulation of glutathione metabolism proteins, features that are typical of ferroptotic cell death [138]. Inactivation of the GPX4, a ferroptosis regulator to alleviate lipid peroxidation, triggers acute renal failure in mice suggesting that genetic mutation to modulate lipid peroxidation plays an important role in the pathological condition [139]. A study reported that propofol could protect against sepsis-induced liver dysfunction through suppressing hepatic oxidative stress and lipid peroxidation [140]. Carnosine, an endogenous histidyl dipeptides, protects cardiac myocytes against lipid peroxidation products of HNE and acrolein toxicity by directly reacting with these aldehydes [141].
As lipid peroxidation promotes the development and progression of critical illness, scientist tried to find strategies to treat/prevent it. However, in the current clinical practice, there is a lack of standardized preventive measures. As regards interventional measures against lipid peroxidation damage in critically ill patients, the interest has been focused on phospholipase A2 (PLA2), cyclooxygenase, and lipoxygenases which is an enzyme involved in the formation of lipid peroxidation [142]. The supplementation of antioxidants in critically ill patients such as vitamin C and E and selenium separately has been found to improve survival and prevent progressive organ dysfunction [143, 144]. As for complex pathological reasons or limited test method, clinical trials of these agents against lipid peroxidation in critical illness have partially failed [145, 146]. As regards for future perspective, detecting techniques targeted to lipid peroxidation biomarkers have been developed. The optimization of understanding the mechanisms of lipid peroxidation has gained a lot of interest, as well as the enhancement of their clinical intervention.
6. Summary and Perspectives
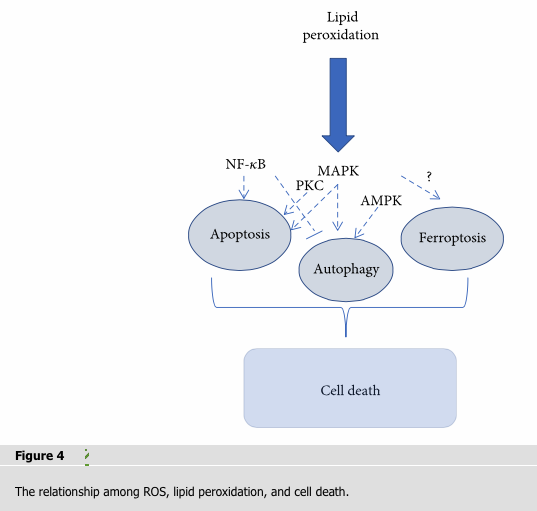
The connectivity of ROS-induced lipid peroxidation caused by apoptosis, autophagy, and ferroptosis manifests themselves in a seamless balance between life and death in response to cellular stress (Figure 4). The generation and elimination of ROS maintain the delicate balance, and the imbalance is associated with various pathologies such as cell proliferation, differentiation, and death. The general endogenous antioxidant system consisting of enzymatic antioxidants (SOD, CAT, GPx, and Trx) and nonenzymatic antioxidants (vitamins or its analogs, minerals, and metabolites) protects cell from oxidative damage. An excess of ROS induces lipid peroxidation via nonenzymatic (iron-dependent) and enzymatic (LOX-catalyzed) pathways which leads to cell death. According to species of lipid peroxidation products, UV/visible spectrophotometry, GC/MS, and ELISA can be used in vivo and C11-BODIPY 581/591 flow cytometry can be applied in vitro for detecting. The products of lipid peroxidation initiate apoptosis in different pathways (NF-κB, MAPK, and PKC) and autophagy by AMPK/mTORC and JNK-Bcl-2/Beclin 1. Ferroptosis is a new form of programmed cell death noticed since 2012 [4]. In recent years, many studies had confirmed that ferroptosis was caused by loss of activity of the GPX4. GPX4 is normally functioned to remove the dangerous products of iron-dependent lipid peroxidation [4, 101, 104]. Although the products of lipid peroxidation have been extensively studied and excessive accumulation of lipid peroxidation has been shown to promote apoptosis and autophagy, their role in ferroptosis is unclear [66, 71, 108]. If the mysteries of lipid peroxidation-induced ferroptosis are solved, new insights and therapeutic strategies for ferroptosis-related human diseases can be provided.
Abbreviations
| ROS: | Reactive oxygen species |
| PUFAs: | Polyunsaturated fatty acids |
| NOXs: | NADPH oxidases |
| XO: | Xanthine oxidase |
| mETC: | Mitochondrial electron-transport chain |
| SOD: | Superoxide dismutase |
| CAT: | Catalase |
| HNE: | Hydroxynonenal |
| PL: | Phospholipid |
| LOX: | Lipoxygenase |
| Alox: | Arachidonate lipoxygenase |
| PLA2: | Phospholipase A2 |
| GPx: | Glutathione peroxidase |
| Trx: | Thioredoxin |
| TrxR: | Thioredoxin reductase |
| GSH: | Glutathione |
| GSSG: | Glutathione disulfide |
| CoQ10: | Coenzyme Q10 |
| HIF-1α: | Hypoxia inducible factor-1α |
| MAPKs: | Mitogen-activated protein kinases |
| ERK: | Extracellular signal-regulated kinase |
| JNK: | Jun N-terminal kinase |
| PKC: | Protein kinase C |
| CMA: | Chaperone-mediated autophagy |
| LC3: | Light chain 3 |
| mTOR: | Mammalian target of rapamycin |
| AMPK: | Adenosine monophosphate-activated protein kinase |
| TSC: | Tuberous sclerosis complex |
| LIP: | Labile iron pool |
| DFO: | Deferoxamine |
| CPX: | Ciclopirox. |
Conflicts of Interest
None of the other authors declared any conflict of interest in this work.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81772046, No. 81560131), the Hubei Province Technology and Innovation Project (Foreign Collaboration of Science and Technology) (No. 2017AHB044), and the Health Commission of Hubei Province (No. WJ2017Z008).
References
- C. A. Ferreira, D. Ni, Z. T. Rosenkrans, and W. Cai, “Scavenging of reactive oxygen and nitrogen species with nanomaterials,” Nano Research, vol. 11, no. 10, pp. 4955–4984, 2018.View at: Publisher Site | Google Scholar
- C. A. K. Lundgren, D. Sjöstrand, O. Biner et al., “Scavenging of superoxide by a membrane-bound superoxide oxidase,” Nature Chemical Biology, vol. 14, no. 8, pp. 788–793, 2018.View at: Publisher Site | Google Scholar
- X. Que, M. Y. Hung, C. Yeang et al., “Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice,” Nature, vol. 558, no. 7709, pp. 301–306, 2018.View at: Publisher Site | Google Scholar
- S. J. Dixon, K. M. Lemberg, M. R. Lamprecht et al., “Ferroptosis: an iron-dependent form of nonapoptotic cell death,” Cell, vol. 149, no. 5, pp. 1060–1072, 2012.View at: Publisher Site | Google Scholar
- G. K. Sakellariou, M. J. Jackson, and A. Vasilaki, “Redefining the major contributors to superoxide production in contracting skeletal muscle. The role of NAD(P)H oxidases,” Free Radical Research, vol. 48, no. 1, pp. 12–29, 2014.View at: Publisher Site | Google Scholar
- G. Guerriero, S. Trocchia, F. K. Abdel-Gawad, and G. Ciarcia, “Roles of reactive oxygen species in the spermatogenesis regulation,” Frontiers in Endocrinology, vol. 5, p. 56, 2014.View at: Publisher Site | Google Scholar
- X. Wen, J. Wu, F. Wang, B. Liu, C. Huang, and Y. Wei, “Deconvoluting the role of reactive oxygen species and autophagy in human diseases,” Free Radical Biology and Medicine, vol. 65, pp. 402–410, 2013.View at: Publisher Site | Google Scholar
- S. Doll and M. Conrad, “Iron and ferroptosis: a still ill-defined liaison,” IUBMB Life, vol. 69, no. 6, pp. 423–434, 2017.View at: Publisher Site | Google Scholar
- M. Kruszewski, “Labile iron pool: the main determinant of cellular response to oxidative stress,” Mutation Research, vol. 531, no. 1-2, pp. 81–92, 2003.View at: Publisher Site | Google Scholar
- G. O. Latunde-Dada, “Ferroptosis: role of lipid peroxidation, iron and ferritinophagy,” Biochimica et Biophysica Acta, vol. 1861, no. 8, pp. 1893–1900, 2017.View at: Publisher Site | Google Scholar
- L. He, T. He, S. Farrar, L. Ji, T. Liu, and X. Ma, “Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species,” Cellular Physiology and Biochemistry, vol. 44, no. 2, pp. 532–553, 2017.View at: Publisher Site | Google Scholar
- Y. Wang, R. Branicky, A. Noe, and S. Hekimi, “Superoxide dismutases: dual roles in controlling ROS damage and regulating ROS signaling,” The Journal of Cell Biology, vol. 217, no. 6, pp. 1915–1928, 2018.View at: Publisher Site | Google Scholar
- S. Li, X. Yang, Z. Feng, P. Wang, W. Zhu, and S. Cui, “Catalase enhances viability of human chondrocytes in culture by reducing reactive oxygen species and counteracting tumor necrosis factor-α-induced apoptosis,” Cellular Physiology and Biochemistry, vol. 49, no. 6, pp. 2427–2442, 2018.View at: Publisher Site | Google Scholar
- E. Zamponi, N. Zamponi, P. Coskun et al., “Nrf2 stabilization prevents critical oxidative damage in Down syndrome cells,” Aging Cell, vol. 17, no. 5, article e12812, 2018.View at: Publisher Site | Google Scholar
- P. Matoušková, B. Hanousková, and L. Skálová, “MicroRNAs as potential regulators of glutathione peroxidases expression and their role in obesity and related pathologies,” International Journal of Molecular Sciences, vol. 19, no. 4, article 1199, 2018.View at: Publisher Site | Google Scholar
- K. Bela, E. Horváth, Á. Gallé, L. Szabados, I. Tari, and J. Csiszár, “Plant glutathione peroxidases: emerging role of the antioxidant enzymes in plant development and stress responses,” Journal of Plant Physiology, vol. 176, pp. 192–201, 2015.View at: Publisher Site | Google Scholar
- L. Halušková, K. Valentovičová, J. Huttová, I. Mistrík, and L. Tamás, “Effect of abiotic stresses on glutathione peroxidase and glutathione S-transferase activity in barley root tips,” Plant Physiology and Biochemistry, vol. 47, no. 11-12, pp. 1069–1074, 2009.View at: Publisher Site | Google Scholar
- A. A. Tinkov, G. Bjørklund, A. V. Skalny et al., “The role of the thioredoxin/thioredoxin reductase system in the metabolic syndrome: towards a possible prognostic marker?” Cellular and Molecular Life Sciences, vol. 75, no. 9, pp. 1567–1586, 2018.View at: Publisher Site | Google Scholar
- J. R. Prigge, L. Coppo, S. S. Martin et al., “Hepatocyte hyperproliferation upon liver-specific co-disruption of thioredoxin-1, thioredoxin reductase-1, and glutathione reductase,” Cell Reports, vol. 19, no. 13, pp. 2771–2781, 2017.View at: Publisher Site | Google Scholar
- M. Rozanowska, A. Cantrell, R. Edge, E. J. Land, T. Sarna, and T. G. Truscott, “Pulse radiolysis study of the interaction of retinoids with peroxyl radicals,” Free Radical Biology and Medicine, vol. 39, no. 10, pp. 1399–1405, 2005.View at: Publisher Site | Google Scholar
- T. Grenier-Larouche, A. Galinier, L. Casteilla, A. C. Carpentier, and A. Tchernof, “Omental adipocyte hypertrophy relates to coenzyme Q10 redox state and lipid peroxidation in obese women,” Journal of Lipid Research, vol. 56, no. 10, pp. 1985–1992, 2015.View at: Publisher Site | Google Scholar
- P. Bullón, L. Román-Malo, F. Marín-Aguilar et al., “Lipophilic antioxidants prevent lipopolysaccharide-induced mitochondrial dysfunction through mitochondrial biogenesis improvement,” Pharmacological Research, vol. 91, pp. 1–8, 2015.View at: Publisher Site | Google Scholar
- A. I. R. N. A. Barros, F. M. Nunes, B. Gonçalves, R. N. Bennett, and A. P. Silva, “Effect of cooking on total vitamin C contents and antioxidant activity of sweet chestnuts (Castanea sativa Mill.),” Food Chemistry, vol. 128, no. 1, pp. 165–172, 2011.View at: Publisher Site | Google Scholar
- G. Y. Lee and S. N. Han, “The role of vitamin E in immunity,” Nutrients, vol. 10, no. 11, p. 1614, 2018.View at: Publisher Site | Google Scholar
- M. Kawser Hossain, A. Abdal Dayem, J. Han et al., “Molecular mechanisms of the anti-obesity and anti-diabetic properties of flavonoids,” International Journal of Molecular Sciences, vol. 17, no. 4, p. 569, 2016.View at: Publisher Site | Google Scholar
- D. D. Marreiro, K. Cruz, J. Morais, J. Beserra, J. Severo, and A. de Oliveira, “Zinc and oxidative stress: current mechanisms,” Antioxidants, vol. 6, no. 2, p. 24, 2017.View at: Publisher Site | Google Scholar
- G. Idelman, D. L. H. Smith, and S. D. Zucker, “Bilirubin inhibits the up-regulation of inducible nitric oxide synthase by scavenging reactive oxygen species generated by the toll-like receptor 4-dependent activation of NADPH oxidase,” Redox Biology, vol. 5, pp. 398–408, 2015.View at: Publisher Site | Google Scholar
- R. Hardeland, “Melatonin and the electron transport chain,” Cellular and Molecular Life Sciences, vol. 74, no. 21, pp. 3883–3896, 2017.View at: Publisher Site | Google Scholar
- J. Blasiak, R. J. Reiter, and K. Kaarniranta, “Melatonin in retinal physiology and pathology: the case of age-related macular degeneration,” Oxidative Medicine and Cellular Longevity, vol. 2016, Article ID 6819736, 12 pages, 2016.View at: Publisher Site | Google Scholar
- M. Xiao, H. Zhong, L. Xia, Y. Tao, and H. Yin, “Pathophysiology of mitochondrial lipid oxidation: role of 4-hydroxynonenal (4-HNE) and other bioactive lipids in mitochondria,” Free Radical Biology and Medicine, vol. 111, pp. 316–327, 2017.View at: Publisher Site | Google Scholar
- G. Spiteller and M. Afzal, “The action of peroxyl radicals, powerful deleterious reagents, explains why neither cholesterol nor saturated fatty acids cause atherogenesis and age-related diseases,” Chemistry - A European Journal, vol. 20, no. 46, pp. 14928–14945, 2014.View at: Publisher Site | Google Scholar
- K. Zarkovic, A. Jakovcevic, and N. Zarkovic, “Contribution of the HNE-immunohistochemistry to modern pathological concepts of major human diseases,” Free Radical Biology and Medicine, vol. 111, pp. 110–126, 2017.View at: Publisher Site | Google Scholar
- W. Łuczaj, A. Gęgotek, and E. Skrzydlewska, “Antioxidants and HNE in redox homeostasis,” Free Radical Biology and Medicine, vol. 111, pp. 87–101, 2017.View at: Publisher Site | Google Scholar
- M. Lal, C. Schöneich, J. Mönig, and K. D. Asmus, “Rate constants for the reactions of halogenated organic radicals,” International Journal of Radiation Biology, vol. 54, no. 5, pp. 773–785, 1988.View at: Publisher Site | Google Scholar
- H. W. Gardner, “Oxygen radical chemistry of polyunsaturated fatty acids,” Free Radical Biology and Medicine, vol. 7, no. 1, pp. 65–86, 1989.View at: Publisher Site | Google Scholar
- S. Bang, S. Park, Y. M. Lee, S. Hong, K. B. Cho, and W. Nam, “Demonstration of the heterolytic O-O bond cleavage of putative nonheme iron(II)-OOH(R) complexes for Fenton and enzymatic reactions,” Angewandte Chemie, vol. 53, no. 30, pp. 7843–7847, 2014.View at: Publisher Site | Google Scholar
- M. C. Michalski, C. Calzada, A. Makino, S. Michaud, and M. Guichardant, “Oxidation products of polyunsaturated fatty acids in infant formulas compared to human milk--a preliminary study,” Molecular Nutrition and Food Research, vol. 52, no. 12, pp. 1478–1485, 2008.View at: Publisher Site | Google Scholar
- D. A. Pratt, K. A. Tallman, and N. A. Porter, “Free radical oxidation of polyunsaturated lipids: new mechanistic insights and the development of peroxyl radical clocks,” Accounts of Chemical Research, vol. 44, no. 6, pp. 458–467, 2011.View at: Publisher Site | Google Scholar
- V. E. Kagan, G. Mao, F. Qu et al., “Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis,” Nature Chemical Biology, vol. 13, no. 1, pp. 81–90, 2017.View at: Publisher Site | Google Scholar
- H. Kuhn, S. Banthiya, and K. van Leyen, “Mammalian lipoxygenases and their biological relevance,” Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, vol. 1851, no. 4, pp. 308–330, 2015.View at: Publisher Site | Google Scholar
- H. Kuhn, J. Belkner, R. Wiesner, and A. R. Brash, “Oxygenation of biological membranes by the pure reticulocyte lipoxygenase,” The Journal of Biological Chemistry, vol. 265, no. 30, pp. 18351–18361, 1990.View at: Google Scholar
- L. T. Morgan, C. P. Thomas, H. Kuhn, and V. B. O'Donnell, “Thrombin-activated human platelets acutely generate oxidized docosahexaenoic-acid-containing phospholipids via 12-lipoxygenase,” The Biochemical Journal, vol. 431, no. 1, pp. 141–148, 2010.View at: Publisher Site | Google Scholar
- S. R. Clark, C. J. Guy, M. J. Scurr et al., “Esterified eicosanoids are acutely generated by 5-lipoxygenase in primary human neutrophils and in human and murine infection,” Blood, vol. 117, no. 6, pp. 2033–2043, 2011.View at: Publisher Site | Google Scholar
- E. Niki, “Lipid peroxidation: physiological levels and dual biological effects,” Free Radical Biology and Medicine, vol. 47, no. 5, pp. 469–484, 2009.View at: Publisher Site | Google Scholar
- J. R. Zhang, A. R. Cazers, B. S. Lutzke, and E. D. Hall, “HPLC-chemiluminescence and thermospray LC/MS study of hydroperoxides generated from phosphatidylcholine,” Free Radical Biology and Medicine, vol. 18, no. 1, pp. 1–10, 1995.View at: Publisher Site | Google Scholar
- A. Ayala, M. F. Munoz, and S. Arguelles, “Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal,” Oxidative Medicine and Cellular Longevity, vol. 2014, Article ID 360438, 31 pages, 2014.View at: Publisher Site | Google Scholar
- E. Niki, “Biomarkers of lipid peroxidation in clinical material,” Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1840, no. 2, pp. 809–817, 2014.View at: Publisher Site | Google Scholar
- Y. Y. Lee, J. M. Galano, C. Oger et al., “Assessment of isoprostanes in human plasma: technical considerations and the use of mass spectrometry,” Lipids, vol. 51, no. 11, pp. 1217–1229, 2016.View at: Publisher Site | Google Scholar
- E. D. Hall and P. K. Andrus, “Measurement of oxygen radicals and lipid peroxidation in neural tissues,” Current Protocols in Neuroscience, vol. 11, pp. 7.17.1–7.17.35, 2001.View at: Publisher Site | Google Scholar
- G. Cheloni and V. I. Slaveykova, “Optimization of the C11-BODIPY581/591 dye for the determination of lipid oxidation in Chlamydomonas reinhardtii by flow cytometry,” Cytometry Part A, vol. 83, pp. 952–961, 2013.View at: Publisher Site | Google Scholar
- Z. Zakeri, W. Bursch, M. Tenniswood, and R. A. Lockshin, “Cell death: programmed, apoptosis, necrosis, or other?” Cell Death & Differentiation, vol. 2, no. 2, pp. 87–96, 1995.View at: Google Scholar
- S. Elmore, “Apoptosis: a review of programmed cell death,” Toxicologic Pathology, vol. 35, no. 4, pp. 495–516, 2007.View at: Publisher Site | Google Scholar
- A. Romero, B. Novoa, and A. Figueras, “The complexity of apoptotic cell death in mollusks: an update,” Fish & Shellfish Immunology, vol. 46, no. 1, pp. 79–87, 2015.View at: Publisher Site | Google Scholar
- A. Ashkenazi and V. M. Dixit, “Death receptors: signaling and modulation,” Science, vol. 281, no. 5381, pp. 1305–1308, 1998.View at: Publisher Site | Google Scholar
- H. Wajant, “The Fas signaling pathway: more than a paradigm,” Science, vol. 296, no. 5573, pp. 1635-1636, 2002.View at: Publisher Site | Google Scholar
- D. R. Green and F. Llambi, “Cell death signaling,” Cold Spring Harbor Perspectives in Biology, vol. 7, no. 12, 2015.View at: Publisher Site | Google Scholar
- Q. Bao and Y. Shi, “Apoptosome: a platform for the activation of initiator caspases,” Cell Death & Differentiation, vol. 14, no. 1, pp. 56–65, 2007.View at: Publisher Site | Google Scholar
- S. J. Riedl and G. S. Salvesen, “The apoptosome: signalling platform of cell death,” Nature Reviews Molecular Cell Biology, vol. 8, no. 5, pp. 405–413, 2007.View at: Publisher Site | Google Scholar
- J. A. Trapani and M. J. Smyth, “Functional significance of the perforin/granzyme cell death pathway,” Nature Reviews Immunology, vol. 2, no. 10, pp. 735–747, 2002.View at: Publisher Site | Google Scholar
- S. S. Metkar, B. Wang, M. L. Ebbs et al., “Granzyme B activates procaspase-3 which signals a mitochondrial amplification loop for maximal apoptosis,” The Journal of Cell Biology, vol. 160, no. 6, pp. 875–885, 2003.View at: Publisher Site | Google Scholar
- I. Grodzovski, M. Lichtenstein, H. Galski, and H. Lorberboum-Galski, “IL-2-granzyme A chimeric protein overcomes multidrug resistance (MDR) through a caspase 3-independent apoptotic pathway,” International Journal of Cancer, vol. 128, no. 8, pp. 1966–1980, 2011.View at: Publisher Site | Google Scholar
- E. R. Elkin, S. M. Harris, and R. Loch-Caruso, “Trichloroethylene metabolite S-(1,2-dichlorovinyl)-l-cysteine induces lipid peroxidation-associated apoptosis via the intrinsic and extrinsic apoptosis pathways in a first-trimester placental cell line,” Toxicology and Applied Pharmacology, vol. 338, pp. 30–42, 2018.View at: Publisher Site | Google Scholar
- P. Chaudhary, R. Sharma, A. Sharma et al., “Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signaling,” Biochemistry, vol. 49, no. 29, pp. 6263–6275, 2010.View at: Publisher Site | Google Scholar
- H. Zhong, M. Xiao, K. Zarkovic et al., “Mitochondrial control of apoptosis through modulation of cardiolipin oxidation in hepatocellular carcinoma: a novel link between oxidative stress and cancer,” Free Radical Biology and Medicine, vol. 102, pp. 67–76, 2017.View at: Publisher Site | Google Scholar
- B. Hoesel and J. A. Schmid, “The complexity of NF-κB signaling in inflammation and cancer,” Molecular Cancer, vol. 12, no. 1, p. 86, 2013.View at: Publisher Site | Google Scholar
- S. Page, C. Fischer, B. Baumgartner et al., “4-Hydroxynonenal prevents NF-κB activation and tumor necrosis factor expression by inhibiting IκB phosphorylation and subsequent proteolysis,” Journal of Biological Chemistry, vol. 274, no. 17, pp. 11611–11618, 1999.View at: Publisher Site | Google Scholar
- C. Bodur, O. Kutuk, T. Tezil, and H. Basaga, “Inactivation of Bcl-2 through IκB kinase (IKK)-dependent phosphorylation mediates apoptosis upon exposure to 4-hydroxynonenal (HNE),” Journal of Cellular Physiology, vol. 227, no. 11, pp. 3556–3565, 2012.View at: Publisher Site | Google Scholar
- C. A. Heckman, J. W. Mehew, and L. M. Boxer, “NF-κB activates Bcl-2 expression in t(14;18) lymphoma cells,” Oncogene, vol. 21, no. 24, pp. 3898–3908, 2002.View at: Publisher Site | Google Scholar
- X. Meng and S. Zhang, “MAPK cascades in plant disease resistance signaling,” Annual Review of Phytopathology, vol. 51, no. 1, pp. 245–266, 2013.View at: Publisher Site | Google Scholar
- K. Lee, I. Seo, M. H. Choi, and D. Jeong, “Roles of mitogen-activated protein kinases in osteoclast biology,” International journal of molecular sciences, vol. 19, no. 10, p. 3004, 2018.View at: Publisher Site | Google Scholar
- H. J. Forman, D. A. Dickinson, and K. E. Iles, “HNE--signaling pathways leading to its elimination,” Molecular aspects of medicine., vol. 24, no. 4-5, pp. 189–194, 2003.View at: Publisher Site | Google Scholar
- G. A. Preston, C. S. Zarella, W. F. Pendergraft 3rd et al., “Novel effects of neutrophil-derived proteinase 3 and elastase on the vascular endothelium Involve In Vivo cleavage of NF-κB and proapoptotic changes in jnk, erk, and p38 mapk signaling pathways,” Journal of the American Society of Nephrology, vol. 13, no. 12, pp. 2840–2849, 2002.View at: Publisher Site | Google Scholar
- K. E. McElhanon, C. Bose, R. Sharma, L. Wu, Y. C. Awasthi, and S. P. Singh, “Gsta4 null mouse embryonic fibroblasts exhibit enhanced sensitivity to oxidants: role of 4-hydroxynonenal in oxidant toxicity,” Open journal of apoptosis, vol. 02, no. 01, pp. 1–11, 2013.View at: Publisher Site | Google Scholar
- C. Giorgi, C. Agnoletto, C. Baldini et al., “Redox control of protein kinase C: cell- and disease-specific aspects,” Antioxidants & redox signaling., vol. 13, no. 7, pp. 1051–1085, 2010.View at: Publisher Site | Google Scholar
- J. D. Violin, J. Zhang, R. Y. Tsien, and A. C. Newton, “A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C,” Journal of cell biology, vol. 161, no. 5, pp. 899–909, 2003.View at: Publisher Site | Google Scholar
- E. Chiarpotto, C. Domenicotti, D. Paola et al., “Regulation of rat hepatocyte protein kinase C β isoenzymes by the lipid peroxidation product 4-hydroxy-2,3-nonenal: a signaling pathway to modulate vesicular transport of glycoproteins,” Hepatology, vol. 29, no. 5, pp. 1565–1572, 1999.View at: Publisher Site | Google Scholar
- J. Zhang, C. M. Burrington, S. K. Davenport et al., “PKCδ regulates hepatic triglyceride accumulation and insulin signaling in Leprdb/db mice,” Biochemical and biophysical research communications, vol. 450, no. 4, pp. 1619–1625, 2014.View at: Publisher Site | Google Scholar
- M. Zhao, L. Xia, and G. Q. Chen, “Protein kinase cδ in apoptosis: a brief overview,” Archivum Immunologiae et Therapiae Experimentalis, vol. 60, no. 5, pp. 361–372, 2012.View at: Publisher Site | Google Scholar
- S. Mei, M. Livingston, J. Hao, L. li, C. Mei, and Z. Dong, “Autophagy is activated to protect against endotoxic acute kidney injury,” Scientific Reports, vol. 6, no. 1, article 22171, 2016.View at: Publisher Site | Google Scholar
- R. Scherz-Shouval and Z. Elazar, “ROS, mitochondria and the regulation of autophagy,” Trends in cell biology, vol. 17, no. 9, pp. 422–427, 2007.View at: Publisher Site | Google Scholar
- N. Mizushima and M. Komatsu, “Autophagy: renovation of cells and tissues,” Cell, vol. 147, no. 4, pp. 728–741, 2011.View at: Publisher Site | Google Scholar
- N. Mizushima, T. Yoshimori, and B. Levine, “Methods in mammalian autophagy research,” Cell, vol. 140, no. 3, pp. 313–326, 2010.View at: Publisher Site | Google Scholar
- M. B. E. Schaaf, T. G. Keulers, M. A. Vooijs, and K. M. A. Rouschop, “LC3/GABARAP family proteins: autophagy-(un)related functions,” The FASEB journal, vol. 30, no. 12, pp. 3961–3978, 2016.View at: Publisher Site | Google Scholar
- L. E. Drake, M. Z. Springer, L. P. Poole, C. J. Kim, and K. F. Macleod, “Expanding perspectives on the significance of mitophagy in cancer,” Seminars in Cancer Biology, vol. 47, pp. 110–124, 2017.View at: Publisher Site | Google Scholar
- J. H. Um and J. Yun, “Emerging role of mitophagy in human diseases and physiology,” BMB Reports, vol. 50, no. 6, pp. 299–307, 2017.View at: Publisher Site | Google Scholar
- J. P. Bernardini, M. Lazarou, and G. Dewson, “Parkin and mitophagy in cancer,” Oncogene, vol. 36, no. 10, pp. 1315–1327, 2017.View at: Publisher Site | Google Scholar
- X. Bian, T. Teng, H. Zhao et al., “Zinc prevents mitochondrial superoxide generation by inducing mitophagy in the setting of hypoxia/reoxygenation in cardiac cells,” Free Radical Research, vol. 52, no. 1, pp. 80–91, 2018.View at: Publisher Site | Google Scholar
- E. F. Fang, Y. Hou, K. Palikaras et al., “Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease,” Nature Neuroscience, vol. 22, no. 3, pp. 401–412, 2019.View at: Publisher Site | Google Scholar
- Y.-F. Chen, H. Liu, X.-J. Luo et al., “The roles of reactive oxygen species (ROS) and autophagy in the survival and death of leukemia cells,” Critical Reviews in Oncology/Hematology, vol. 112, pp. 21–30, 2017.View at: Publisher Site | Google Scholar
- M. Dodson, W. Y. Wani, M. Redmann et al., “Regulation of autophagy, mitochondrial dynamics, and cellular bioenergetics by 4-hydroxynonenal in primary neurons,” Autophagy, vol. 13, no. 11, pp. 1828–1840, 2017.View at: Publisher Site | Google Scholar
- H. Ma, R. Guo, L. Yu, Y. Zhang, and J. Ren, “Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde,” European Heart Journal, vol. 32, no. 8, pp. 1025–1038, 2011.View at: Publisher Site | Google Scholar
- T. U. Krohne, E. Kaemmerer, F. G. Holz, and J. Kopitz, “Lipid peroxidation products reduce lysosomal protease activities in human retinal pigment epithelial cells via two different mechanisms of action,” Experimental Eye Research, vol. 90, no. 2, pp. 261–266, 2010.View at: Publisher Site | Google Scholar
- A. Eisenberg-Lerner and A. Kimchi, “The paradox of autophagy and its implication in cancer etiology and therapy,” Apoptosis, vol. 14, no. 4, pp. 376–391, 2009.View at: Publisher Site | Google Scholar
- E. A. Dunlop and A. R. Tee, “mTOR and autophagy: a dynamic relationship governed by nutrients and energy,” Seminars in Cell & Developmental Biology, vol. 36, pp. 121–129, 2014.View at: Publisher Site | Google Scholar
- M. Ito, T. Yurube, K. Kakutani et al., “Selective interference of mTORC1/RAPTOR protects against human disc cellular apoptosis, senescence, and extracellular matrix catabolism with Akt and autophagy induction,” Osteoarthritis and Cartilage, vol. 25, no. 12, pp. 2134–2146, 2017.View at: Publisher Site | Google Scholar
- V. W. Dolinsky, A. Y. M. Chan, I. Robillard Frayne, P. E. Light, C. Des Rosiers, and J. R. B. Dyck, “Resveratrol prevents the prohypertrophic effects of oxidative stress on LKB1,” Circulation, vol. 119, no. 12, pp. 1643–1652, 2009.View at: Publisher Site | Google Scholar
- D. Yan, G. An, and M. T. Kuo, “C-Jun N-terminal kinase signalling pathway in response to cisplatin,” Journal of Cellular and Molecular Medicine, vol. 20, no. 11, pp. 2013–2019, 2016.View at: Publisher Site | Google Scholar
- C. He, Y. Wei, K. Sun et al., “Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism,” Cell, vol. 154, no. 5, pp. 1085–1099, 2013.View at: Publisher Site | Google Scholar
- S. R. Klein, S. Piya, Z. Lu et al., “C-Jun N-terminal kinases are required for oncolytic adenovirus-mediated autophagy,” Oncogene, vol. 34, no. 41, pp. 5295–5301, 2015.View at: Publisher Site | Google Scholar
- P. Haberzettl and B. G. Hill, “Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response,” Redox Biology, vol. 1, no. 1, pp. 56–64, 2013.View at: Publisher Site | Google Scholar
- B. R. Stockwell, J. P. Friedmann Angeli, H. Bayir et al., “Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease,” Cell, vol. 171, no. 2, pp. 273–285, 2017.View at: Publisher Site | Google Scholar
- S. Masaldan, S. A. S. Clatworthy, C. Gamell et al., “Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis,” Redox Biology, vol. 14, pp. 100–115, 2018.View at: Publisher Site | Google Scholar
- R. Schreiber, B. Buchholz, A. Kraus et al., “Lipid peroxidation drives renal cyst growth in vitro through activation of TMEM16A,” Journal of the American Society of Nephrology, vol. 30, no. 2, pp. 228–242, 2019.View at: Publisher Site | Google Scholar
- M. Conrad, V. E. Kagan, H. Bayir et al., “Regulation of lipid peroxidation and ferroptosis in diverse species,” Genes & Development, vol. 32, no. 9-10, pp. 602–619, 2018.View at: Publisher Site | Google Scholar
- T. M. Seibt, B. Proneth, and M. Conrad, “Role of GPX4 in ferroptosis and its pharmacological implication,” Free Radical Biology and Medicine, vol. 133, pp. 144–152, 2019.View at: Publisher Site | Google Scholar
- D. A. Stoyanovsky, Y. Y. Tyurina, I. Shrivastava et al., “Iron catalysis of lipid peroxidation in ferroptosis: regulated enzymatic or random free radical reaction?” Free Radical Biology and Medicine, vol. 133, pp. 153–161, 2019.View at: Publisher Site | Google Scholar
- S. Masaldan, A. I. Bush, D. Devos, A. S. Rolland, and C. Moreau, “Striking while the iron is hot: iron metabolism and ferroptosis in neurodegeneration,” Free Radical Biology and Medicine, vol. 133, pp. 221–233, 2019.View at: Publisher Site | Google Scholar
- H. Feng and B. R. Stockwell, “Unsolved mysteries: how does lipid peroxidation cause ferroptosis?” PLoS Biology, vol. 16, no. 5, article e2006203, 2018.View at: Publisher Site | Google Scholar
- V. Ribas, C. GarcÃa-Ruiz, and J. Ã.©. C. Fernández-Checa, “Glutathione and mitochondria,” Frontiers in Pharmacology, vol. 5, p. 151, 2014.View at: Publisher Site | Google Scholar
- A. Jelinek, L. Heyder, M. Daude et al., “Mitochondrial rescue prevents glutathione peroxidase-dependent ferroptosis,” Free Radical Biology and Medicine, vol. 117, pp. 45–57, 2018.View at: Publisher Site | Google Scholar
- Y. Kinowaki, M. Kurata, S. Ishibashi et al., “Glutathione peroxidase 4 overexpression inhibits ROS-induced cell death in diffuse large B-cell lymphoma,” Laboratory Investigation, vol. 98, no. 5, pp. 609–619, 2018.View at: Publisher Site | Google Scholar
- W. S. Yang, R. SriRamaratnam, M. E. Welsch et al., “Regulation of ferroptotic cancer cell death by GPX4,” Cell, vol. 156, no. 1-2, pp. 317–331, 2014.View at: Publisher Site | Google Scholar
- W. S. Yang and B. R. Stockwell, “Ferroptosis: death by lipid peroxidation,” Trends in Cell Biology, vol. 26, no. 3, pp. 165–176, 2016.View at: Publisher Site | Google Scholar
- D. Shin, E. H. Kim, J. Lee, and J. L. Roh, “Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer,” Free Radical Biology and Medicine, vol. 129, pp. 454–462, 2018.View at: Publisher Site | Google Scholar
- J. P. Friedmann Angeli and M. Conrad, “Selenium and GPX4, a vital symbiosis,” Free Radical Biology and Medicine, vol. 127, pp. 153–159, 2018.View at: Publisher Site | Google Scholar
- I. Ingold and M. Conrad, “Selenium and iron, two elemental rivals in the ferroptotic death process,” Oncotarget, vol. 9, no. 32, pp. 22241-22242, 2018.View at: Publisher Site | Google Scholar
- D. R. Green, “An element of life,” Cell, vol. 172, no. 3, pp. 389-390, 2018.View at: Publisher Site | Google Scholar
- K. Shimada, R. Skouta, A. Kaplan et al., “Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis,” Nature Chemical Biology, vol. 12, no. 7, pp. 497–503, 2016.View at: Publisher Site | Google Scholar
- W. B. Musgrave, H. Yi, D. Kline et al., “Probing the origins of glutathione biosynthesis through biochemical analysis of glutamate-cysteine ligase and glutathione synthetase from a model photosynthetic prokaryote,” Biochemical Journal, vol. 450, no. 1, pp. 63–72, 2013.View at: Publisher Site | Google Scholar
- P. Koppula, Y. Zhang, L. Zhuang, and B. Gan, “Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer,” Cancer Communications, vol. 38, no. 1, p. 12, 2018.View at: Publisher Site | Google Scholar
- S. J. Dixon, D. N. Patel, M. Welsch et al., “Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis,” eLife, vol. 3, article e02523, 2014.View at: Publisher Site | Google Scholar
- Z. Firtina Karagonlar, D. Koc, E. Iscan, E. Erdal, and N. Atabey, “Elevated hepatocyte growth factor expression as an autocrine c-Met activation mechanism in acquired resistance to sorafenib in hepatocellular carcinoma cells,” Cancer Science, vol. 107, no. 4, pp. 407–416, 2016.View at: Publisher Site | Google Scholar
- E. Lachaier, C. Louandre, C. Godin et al., “Sorafenib induces ferroptosis in human cancer cell lines originating from different solid tumors,” Anticancer Research, vol. 34, no. 11, pp. 6417–6422, 2014.View at: Google Scholar
- E. H. Kim, D. Shin, J. Lee, A. R. Jung, and J. L. Roh, “CISD2 inhibition overcomes resistance to sulfasalazine-induced ferroptotic cell death in head and neck cancer,” Cancer Letters, vol. 432, pp. 180–190, 2018.View at: Publisher Site | Google Scholar
- P. Nagakannan, M. I. Islam, S. Karimi-Abdolrezaee, and E. Eftekharpour, “Inhibition of VDAC1 protects against glutamate-induced oxytosis and mitochondrial fragmentation in hippocampal HT22 cells,” Cellular and Molecular Neurobiology, vol. 39, no. 1, pp. 73–85, 2019.View at: Publisher Site | Google Scholar
- K. Wang, X. Zhu, K. Zhang et al., “Neuroprotective effect of puerarin on glutamate-induced cytotoxicity in differentiated Y-79 cells via inhibition of ROS generation and Ca2+ influx,” International journal of molecular sciences, vol. 17, no. 7, p. 1109, 2016.View at: Publisher Site | Google Scholar
- J. Lewerenz, M. Klein, and A. Methner, “Cooperative action of glutamate transporters and cystine/glutamate antiporter system Xc- protects from oxidative glutamate toxicity,” Journal of Neurochemistry, vol. 98, no. 3, pp. 916–925, 2006.View at: Publisher Site | Google Scholar
- S. Nishizawa, H. Araki, Y. Ishikawa et al., “Low tumor glutathione level as a sensitivity marker for glutamate‑cysteine ligase inhibitors,” Oncology Letters, vol. 15, no. 6, pp. 8735–8743, 2018.View at: Publisher Site | Google Scholar
- J. Zhang and X. Chen, “p53 tumor suppressor and iron homeostasis,” The FEBS Journal, vol. 286, no. 4, pp. 620–629, 2018.View at: Publisher Site | Google Scholar
- Y. Y. Yien, J. Shi, C. Chen et al., “FAM210B is an erythropoietin target and regulates erythroid heme synthesis by controlling mitochondrial iron import and ferrochelatase activity,” Journal of Biological Chemistry, vol. 293, no. 51, pp. 19797–19811, 2018.View at: Publisher Site | Google Scholar
- F. Zohora, K. Bidad, Z. Pourpak, and M. Moin, “Biological and immunological aspects of iron deficiency anemia in cancer development: a narrative review,” Nutrition and Cancer, vol. 70, no. 4, pp. 546–556, 2018.View at: Publisher Site | Google Scholar
- I. Silva, V. Rausch, T. Peccerella, G. Millonig, H. K. Seitz, and S. Mueller, “Hypoxia enhances H2O2-mediated upregulation of hepcidin: evidence for NOX4-mediated iron regulation,” Redox Biology, vol. 16, pp. 1–10, 2018.View at: Publisher Site | Google Scholar
- O. Stehling, A. D. Sheftel, and R. Lill, “Chapter 12 twelve controlled expression of iron‐sulfur cluster assembly components for respiratory chain complexes in mammalian cells,” Methods in Enzymology, vol. 456, pp. 209–231, 2009.View at: Publisher Site | Google Scholar
- J. D. Mancias, X. Wang, S. P. Gygi, J. W. Harper, and A. C. Kimmelman, “Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy,” Nature, vol. 509, no. 7498, pp. 105–109, 2014.View at: Publisher Site | Google Scholar
- D. M. Ward and J. Kaplan, “Ferroportin-mediated iron transport: expression and regulation,” Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1823, no. 9, pp. 1426–1433, 2012.View at: Publisher Site | Google Scholar
- M. Krijt, A. Jirkovska, T. Kabickova, V. Melenovsky, J. Petrak, and D. Vyoral, “Detection and quantitation of iron in ferritin, transferrin and labile iron pool (LIP) in cardiomyocytes using 55Fe and storage phosphorimaging,” Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1862, no. 12, pp. 2895–2901, 2018.View at: Publisher Site | Google Scholar
- H. Lv and P. Shang, “The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes,” Metallomics, vol. 10, no. 7, pp. 899–916, 2018.View at: Publisher Site | Google Scholar
- D. Martin-Sanchez, O. Ruiz-Andres, J. Poveda et al., “Ferroptosis, but not necroptosis, is important in nephrotoxic folic acid–induced AKI,” Journal of the American Society of Nephrology, vol. 28, no. 1, pp. 218–229, 2017.View at: Publisher Site | Google Scholar
- J. P. Friedmann Angeli, M. Schneider, B. Proneth et al., “Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice,” Nature Cell Biology, vol. 16, no. 12, pp. 1180–1191, 2014.View at: Publisher Site | Google Scholar
- G. J. Wu, Y. W. Lin, H. C. Tsai, Y. W. Lee, J. T. Chen, and R. M. Chen, “Sepsis-induced liver dysfunction was ameliorated by propofol via suppressing hepatic lipid peroxidation, inflammation, and drug interactions,” Life Sciences, vol. 213, pp. 279–286, 2018.View at: Publisher Site | Google Scholar
- J. Zhao, D. K. Posa, V. Kumar et al., “Carnosine protects cardiac myocytes against lipid peroxidation products,” Amino Acids, vol. 51, no. 1, pp. 123–138, 2019.View at: Publisher Site | Google Scholar
- T. S. Anthonymuthu, E. M. Kenny, and H. Bayir, “Therapies targeting lipid peroxidation in traumatic brain injury,” Brain Research, vol. 1640, Part A, pp. 57–76, 2016.View at: Publisher Site | Google Scholar
- P. E. Marik, V. Khangoora, R. Rivera, M. H. Hooper, and J. Catravas, “Hydrocortisone, vitamin C, and thiamine for the treatment of severe sepsis and septic shock,” Chest, vol. 151, no. 6, pp. 1229–1238, 2017.View at: Publisher Site | Google Scholar
- J. B. Belsky, C. R. Wira, V. Jacob, J. E. Sather, and P. J. Lee, “A review of micronutrients in sepsis: the role of thiamine, l-carnitine, vitamin C, selenium and vitamin D,” Nutrition Research Reviews, vol. 31, no. 2, pp. 281–290, 2018.View at: Publisher Site | Google Scholar
- R. G. B. de Oliveira Nascimento Freitas, R. J. N. Nogueira, S. M. F. Cozzolino, A. C. J. Vasques, and G. Hessel, “Influence of selenium supplementation on patients with inflammation: a pilot double blind randomized study,” Nutrition, vol. 41, pp. 32–36, 2017.View at: Publisher Site | Google Scholar
- W. A. C. (. K.). Koekkoek and A. R. H. van Zanten, “Antioxidant vitamins and trace elements in critical illness,” Nutrition in Clinical Practice, vol. 31, no. 4, pp. 457–474, 2016.View at: Publisher Site | Google Scholar