beyond reason
Approximately 1%–1.5% of cellular proteins are catabolized per hour by autophagy, even under nutrient-rich conditions in the liver.
- 음식을 충분히 복용하는 상황에서도 간에서는 대략 시간당 1-1.5%의 세포단백질이 이화되는 과정을 거침.

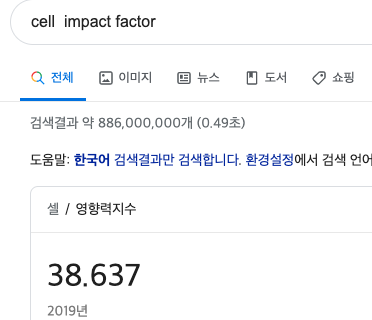
Macroautophagy: A portion of cytoplasm, including organelles, is enclosed by an isolation membrane (also called phagophore) to form an autophagosome. The outer membrane of the autophagosome fuses with the lysosome, and the internal material is degraded in the autolysosome. Microautophagy: Small pieces of the cytoplasm are directly engulfed by inward invagination of the lysosomal or late endosomal membrane. Chaperone-mediated autophagy: Substrate proteins containing a KFERQ-like pentapeptide sequence are first recognized by cytosolic Hsc70 and cochaperones. Then they are translocated into the lysosomal lumen after binding with lysosomal Lamp-2A. After all three types of autophagy, the resultant degradation products can be used for different purposes, such as new protein synthesis, energy production, and gluconeogenesis.
abstract
Autophagy is the major intracellular degradation system by which cytoplasmic materials are delivered to and degraded in the lysosome. However, the purpose of autophagy is not the simple elimination of materials, but instead, autophagy serves as a dynamic recycling system that produces new building blocks and energy for cellular renovation and homeostasis.
Here we provide a multidisciplinary review of our current understanding of autophagy's role in metabolic adaptation, intracellular quality control, and renovation during development and differentiation. We also explore how recent mouse models in combination with advances in human genetics are providing key insights into how the impairment or activation of autophagy contributes to pathogenesis of diverse diseases, from neurodegenerative diseases such as Parkinson disease to inflammatory disorders such as Crohn disease.
Main Text
All living organisms undergo continuous renovation. In humans, cells and intracellular components are constantly remodeled and recycled. This is, in part, in order to replace old components with fresh, better-quality ones. However, when components are replaced with different types, a net change in character results. Such “cellular renovation” requires synthesis of new components but also degradation of pre-existing materials, which can serve as building blocks.
Eukaryotic cells have two major degradation systems, the lysosome and the proteasome. Proteasomal degradation has high selectivity; the proteasome generally recognizes only ubiquitinated substrates, which are primarily short-lived proteins. By contrast, degradation in the lysosome does not follow such a simple pattern. Extracellular material and plasma membrane proteins can be delivered to lysosomes for degradation via the endocytic pathway. Furthermore, cytosolic components and organelles can also be delivered to the lysosome by autophagy (Figure 1).
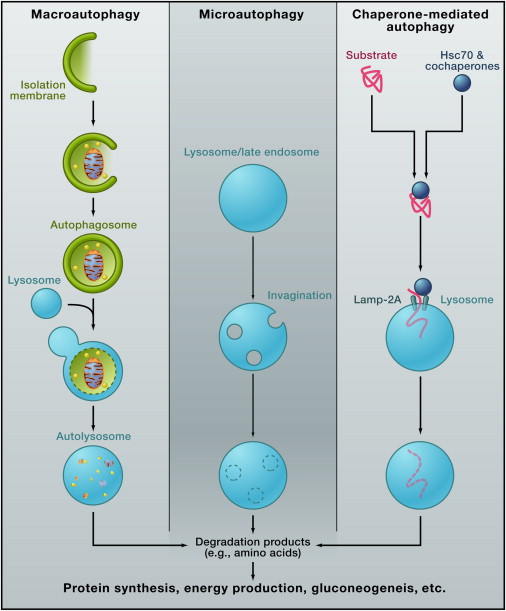
Figure 1Different Types of Autophagy
The lysosome is often described as a “cellular garbage can,” and its more positive roles in cellular renovation, particularly those involving autophagy, have not been well appreciated. In the 1990s, genetic studies in yeast identified a series of autophagy-related (ATG) genes (
,
). The results of these studies greatly increased our understanding of the mechanism and function of autophagy. In particular, analyses of autophagy-defective organisms have revealed numerous physiological and pathological roles of autophagy at both the cellular and whole-organism levels. In this Review, we summarize the current knowledge of autophagy and discuss the multidisciplinary function of autophagy in renovation of the cell and the organism.
Mechanisms of Autophagy
Autophagy is a generic term for all pathways by which cytoplasmic materials are delivered to the lysosome in animal cells or the vacuole in plant and yeast cells. There are roughly three classes of autophagy (Figure 1): macroautophagy, microautophagy, and chaperone-mediated autophagy.
Macroautophagy uses the intermediate organelle “autophagosome.” An isolation membrane (also termed phagophore) sequesters a small portion of the cytoplasm, including soluble materials and organelles, to form the autophagosome. The autophagosome fuses with the lysosome to become an autolysosome and degrade the materials contained within it. Autophagosomes may fuse with endosomes before fusion with lysosomes.
In microautophagy, the lysosome itself engulfs small components of the cytoplasm by inward invagination of the lysosomal membrane (Figure 1).
Membrane dynamics during microautophagy may be quite similar or identical to that of endosomal sorting complex required for transport (ESCRT)-dependent multivesicular body (MVB) formation, which occurs in the late endosome. In fact, significant amounts of cytosolic proteins are incorporated into the endosomal lumen during MVB formation both in bulk and selectively (
).
The third type of autophagy is chaperone-mediated autophagy. This class does not involve membrane reorganization; instead, substrate proteins directly translocate across the lysosomal membrane during chaperone-mediated autophagy (Figure 1). The chaperone protein Hsc70 (heat shock cognate 70) and cochaperones specifically recognize cytosolic proteins that contain a KFERQ-like pentapeptide (
). The transmembrane protein Lamp-2A, which is an isoform of Lamp-2, acts as a receptor on the lysosome, and unfolded proteins are delivered into the lysosomal lumen through a multimeric translocation complex.
Macroautophagy is thought to be the major type of autophagy, and it has been studied most extensively compared to microautophagy and chaperone-mediated autophagy. Therefore, herein we refer to macroautophagy simply as “autophagy.”
Autophagy is highly inducible, with starvation and other stresses rapidly increasing the number of autophagosomes. Autophagosomes are generated on or in close proximity to the endoplasmic reticulum (ER) (Figure 2) (
,
). However, it remains unclear whether the ER membrane is directly used for autophagosome formation. Recent studies suggest that additional membranes derived from the Golgi complex, the mitochondria, and the plasma membrane also contribute to autophagosome formation (
,
,
,
). Thus, autophagosome formation likely involves multiple, complex processes.
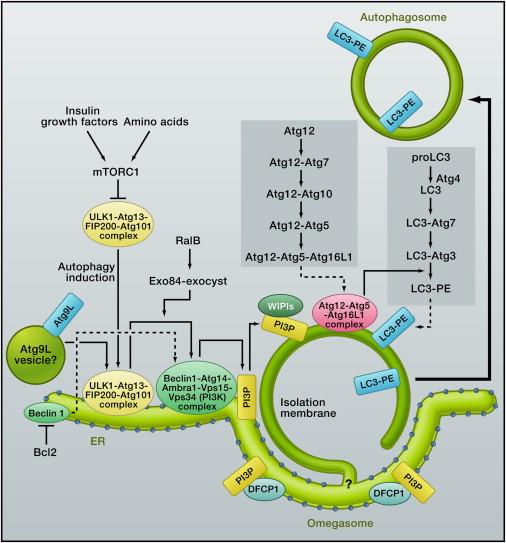
Figure 2Autophagosome Formation and Atg Proteins in Mammalian Cells
Multiple Atg proteins govern autophagosome formation. Among the 35 Atg proteins thus far identified in yeast, Atg1–10, 12–14, 16, and 18 are the “core Atg proteins” (
). These proteins are required for autophagosome formation, in addition to Atg17, 29, and 31. The core Atg proteins are shared by other autophagy-related pathways, such as pexophagy (autophagic degradation of the peroxisome) and the cytoplasm-to-vacuole targeting pathway, which have been discussed in more detail in other reviews (
,
,
).
The core Atg proteins are highly conserved in other eukaryotes, including mammals, and they act in a similar hierarchical manner in yeast and mammals (
,
). Figure 2 summarizes their functional steps in mammalian cells, and more details for this process are described extensively elsewhere (
,
,
).
Adaptive Metabolic Response
The proteasome functions as a major generator of amino acids under normal nutrient-rich conditions, but autophagy's contribution to amino acid production increases when cells are starved (
). Limitation of various types of nutrients, such as amino acids, growth factors, oxygen, and energy, can induce autophagy (
,
). Among these nutrients, starvation of nitrogen or amino acids induces the highest levels of autophagy in yeast and cultured mammalian cells, respectively. This is quite reasonable because the main products of autophagy are amino acids derived from cellular proteins. Restoration of cellular (or local) levels of amino acids reactivates the serine/threonine protein kinase mTORC1 (mammalian target of rapamycin complex 1) and terminates autophagy (
). Autophagy, therefore, constitutes a negative feedback loop in response to nutrient starvation.
When yeast cells are cultured in nitrogen-free medium, autophagy-deficient cells rapidly decrease intracellular amino acid levels (
) and lose their viability (
). Similarly, mice with systemic deletion of Atg3 (
), Atg5 (
), and Atg7 (
) die immediately after birth and show reduced amino acid levels in tissues and plasma during the neonatal starvation period. Thus, enhanced degradation of self-components by autophagy is a critical survival response against starvation conditions.
An important question is, how do cells use these amino acids produced by autophagy (Figure 1, Figure 3)? Initial induction of autophagy is very rapid and occurs before energy fuels are completely exhausted. For instance, mice starved for 24 hr show increased autophagy in many tissues, but they still have sufficient lipids (glycogen may be consumed during the first day). Therefore, it is unlikely that autophagy simply supplies energy in these settings. In fact, several studies have suggested that autophagy-derived amino acids are used to synthesize proteins essential for starvation adaptation.
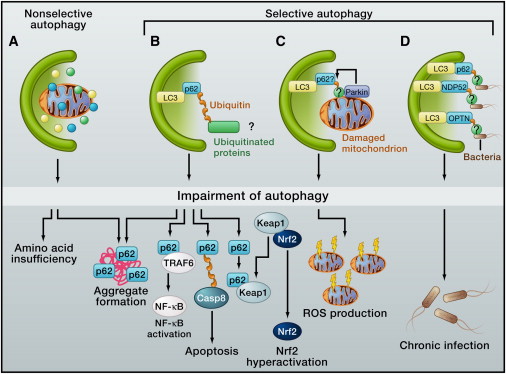
Figure 3 Nxxonselective and Selective Autophagy
Protein anabolism is generally downregulated in starved cells, but synthesis of certain types of proteins continues or is even upregulated during starvation. These include vacuolar/lysosomal enzymes, respiratory chain proteins, antioxidant enzymes, and proteins involved in pathways of amino acid biosynthesis (
,
). Autophagy-deficient yeast cells fail to synthesize these proteins during starvation. As a result, these mutants loose respiratory function and accumulate higher levels of reactive oxygen species, which further decreases their mitochondrial DNA content (
). These appear to be the major mechanisms that rapidly kill autophagy-deficient yeast cells during nitrogen limitation.
Nonetheless, autophagy seems to be an important energy generator in certain settings. Amino acids can be converted into intermediates of the tricarboxylic acid (TCA) cycle and thus contribute to ATP production. Metabolome analysis of Ras-expressing cancer cells shows that autophagy is important for maintenance of TCA cycle metabolites (
). It is interesting that citrate, aconitate, and isocitrate, which are solely produced in mitochondria, are specifically reduced, suggesting that autophagy could be important not only for providing TCA metabolites but also for quality control of mitochondria. In addition, breakdown of lipid droplets by autophagy (lipophagy) may also account for its energy-producing role, especially in the liver (
).
Finally, amino acids are thought to be an important source for gluconeogenesis in the liver. In fact, when Atg7 is deleted specifically in the liver, the mutant mice show reduced levels of blood glucose and amino acids after 24 hr of starvation (
). This finding suggests that amino acids generated inside the liver are used for gluconeogenesis and maintenance of the plasma pool of amino acids. How much autophagy contributes to overall gluconeogenesis is still unknown.
Intracellular Quality Control by Nxxonselective and Selective Autophagy
Approximately 1%–1.5% of cellular proteins are catabolized per hour by autophagy, even under nutrient-rich conditions in the liver. It is unclear how much basal autophagy contributes to macromolecule synthesis and energy production in the steady state by supplying amino acids, glucose, and free fatty acids. Nevertheless, basal autophagy acts as the quality-control machinery for cytoplasmic components, and it is crucial for homeostasis of various postmitotic cells, such as neurons and hepatocytes. Although this quality control could be partially achieved by nxxonselective autophagy, increasing evidence indicates that “selective” autophagy degrades specific proteins, organelles, and invading bacteria (Figure 3). Selective autophagy occurs constitutively and can also be induced in response to cellular stresses.
Selective Degradation of p62
One of the best characterized substrates of selective autophagy is p62, which is also known as sequestosome 1/SQSTM1. p62 is an ubiquitously expressed cellular protein, which is conserved in animals but not in plants and fungi. p62 directly interacts with LC3 (microtubule-associated protein light chain 3) on the isolation membrane through the LC3-interacting region (Figure 3). (LC3 is the mammalian homolog of Atg8 in yeast.) Subsequently, p62 is incorporated into the autophagosome and then degraded (
,
).
Impairment of autophagy is accompanied by accumulation of p62. This leads to the formation of large aggregates, which include p62 and ubiquitin (
). Similar inclusion bodies with p62 and ubiquitin have been identified in various neurodegenerative diseases, including Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis; liver disorders, including alcoholic hepatitis and steatohepatitis; and cancers, including malignant glioma and hepatocellular carcinoma (
). When Atg7 is disrupted in livers and brains of mice, p62-positive aggregates are observed in their hepatocytes and neurons, respectively. Interestingly, these aggregates, as well those in human hepatocellular carcinoma cells, are completely dispersed by the additional loss of p62 (
,
). These findings implicate p62 in the formation of disease-related inclusion bodies (Figure 3).
p62 functions as a signaling hub that may determine whether cells survive by activating the TNF receptor-associated factor 6 (TRAF6)–NF-κB pathway or die by facilitating the aggregation of caspase-8 and downstream effector caspases (
). On the other hand, p62 interacts with the Nrf2-binding site on Keap1, a component of Cullin3-type ubiquitin ligase for Nrf2. This interaction stabilizes Nrf2 and activates the transcription of Nrf2 target genes, including a battery of antioxidant proteins (
,
). It is thus possible that excess accumulation or aggregation of p62 leads to hyperactivation of these signaling pathways (Figure 3).
Selective Degradation of Ubiquitinated Cargos
Almost all tissues with defective autophagy display an accumulation of polyubiquitinated proteins (
). Loss of autophagy is thought to delay global turnover of cytoplasmic components (
) and impair the degradation of substrates destined for the proteasome (
). These effects could partially explain the accumulation of misfolded and unfolded proteins followed by the formation of inclusion bodies.
However, p62 has a ubiquitin-associated (UBA) domain. Thus, it has been proposed that p62 may be an autophagy receptor for degrading ubiquitinated cargos, including ubiquitinated aggregates, damaged mitochondria, ubiquitinated midbody rings, ubiquitin-tagged peroxisomes, ubiquitinated microbes, ribosomal proteins, and virus capsid proteins (
,
) (Figure 3). p62 and other adaptor proteins, such as NDP52 (
) and optineurin (
), mediate the degradation of invading microbes via their interaction with ubiquitination (Figure 3). This selective autophagy could be regulated by posttranslational modification of the adaptors. For instance, TANK-binding kinase (TBK1) phosphorylates optineurin, which enhances its binding affinity to LC3 and thereby suppresses growth of invading microbes (
).
Although a large number of studies suggest that p62 acts as an ubiquitin adaptor, it is still unknown whether soluble ubiquitinated proteins are also degraded through p62 binding. A mass spectrometric analysis clearly demonstrated that ubiquitinated proteins in autophagy-deficient livers and brains do not show any linkage specificity, indicating that specific polyubiquitin chain linkage is not the decisive signal for autophagic degradation (
). The simultaneous knockout of either p62 or Nrf2 completely suppresses the increase in ubiquitin conjugates in Atg7-deficient liver and brain (
). Therefore, the accumulation of ubiquitinated proteins in tissues defective in autophagy might be attributed to p62-mediated activation of Nrf2, resulting in global transcriptional changes to ubiquitin-associated genes. Further studies will be needed to elucidate more precisely the mechanism of degradation of ubiquitinated proteins by autophagy.
Degradation of Damaged Mitochondria: Implications for the Pathogenesis of Parkinson Disease
Recent studies have described the molecular mechanism by which damaged mitochondria are selectively targeted for autophagy, and these studies also suggest that the defects in this process underlie familial Parkinson disease (
). PINK1, a mitochondrial kinase, and Parkin, an E3 ubiquitin ligase, have been genetically linked to both Parkinson disease and a pathway that prevents progressive mitochondrial damage and dysfunction. When mitochondria are damaged and depolarized, PINK1 becomes stabilized and recruits Parkin to the damaged mitochondria (
,
,
,
). Parkin ubiquitinates various mitochondrial outer membrane proteins, which could trigger mitophagy. However, the precise substrate of Parkin, which is essential for mitophagy, is still unknown (Figure 3). Of note, mutations in PINK1 and Parkin, which are associated with Parkinson disease, are known to impair mitophagy (
,
,
,
), suggesting that there is a link between defective mitophagy and Parkinson disease. Accumulation of damaged mitochondria would cause oxidative stress and loss of neuronal cells.
Nevertheless, many questions remain about mitophagy and Parkin. First, it is unknown how the autophagosome recognizes these ubiquitinated mitochondria. Although p62 has been implicated in this recognition process, elimination of mitochondria occurs normally in p62-deficient cells (
,
). p62 seems to be required for the clustering of depolarized mitochondria in the perinuclear region. The role of other mitochondrial adaptor proteins such as Nix remains unknown (
). Second, Parkin appears to function in other degradative processes besides mitophagy. Parkin can induce degradation of a broad range of mitochondrial outer membrane proteins, such as mitofusin 1/2 and Tom20 (
,
,
), which may also be important for mitochondrial quality control. Third, in contrast to in vitro studies, a recent in vivo study showed that depolarized mitochondria do not recruit Parkin in dopamine neurons (
). Thus, Parkin seems to have multiple functions in mitochondrial quality control and neuronal cell survival. More studies are needed to fully understand the physiological and pathological role of this protein.
Renovation during Differentiation and Development
Development and differentiation processes are often accompanied by drastic cellular and tissue remodeling, which requires enhanced degradation. Autophagy can “kill two birds with one stone” during this remodeling; it eliminates pre-existing materials and provides support for the subsequent creation of new components (
). For example, autophagy has been shown to be required for formation of spores in yeast (
) and dauer larvae in Caenorhabditis elegans (
), both of which are triggered by starvation to sustain the organism during adverse conditions. Autophagy also plays a crucial role in insect metamorphosis (
). It is possible that autophagy-derived amino acids can be used as essential building blocks in these processes.
The most dramatic cellular renovation may occur shortly after fertilization. Maternal proteins and messenger RNAs (mRNAs) are extensively degraded while new proteins encoded by the zygotic genome are synthesized. In mammals, fertilization induces massive autophagy, which plays an essential role in early embryogenesis (
) (Figure 4). Nutrient availability may be limited in embryos until implantation, and autophagy functions as a major nutrient-providing system during this period. In addition, autophagy in early embryos is essential for selective elimination of paternal mitochondria in C. elegans. This could be a key mechanism underlying maternal inheritance of mitochondrial DNA and may be conserved in mammals (
,
).
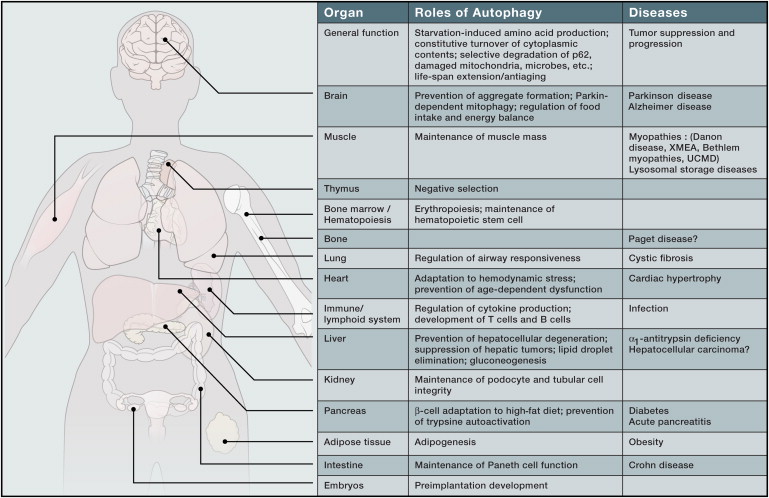
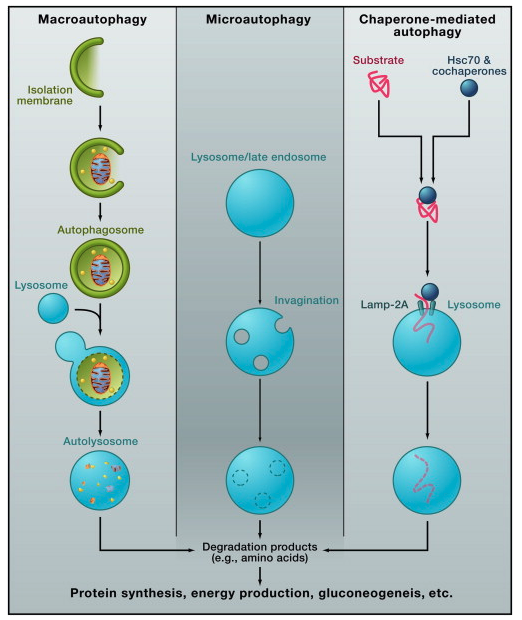
Figure 4 Physiological and Pathological Roles of Autophagy
Atg3−/− (
), Atg5−/− (
), Atg7−/− (
), Atg9−/− (
), and Atg16L1−/− (
) mice die shortly after birth without apparent anatomical abnormalities. However, tissue-specific gene-targeting studies have later revealed that autophagy functions in several specific lineages of differentiation, such as adipocytes, erythrocytes, T cells, and B-1a cells (Figure 4). Autophagy may play an important role in complete or partial elimination of mitochondria during these processes (
). In a separate mechanism, autophagy in thymic epithelial cells could be involved in the establishment of self-tolerance of T cells; specifically, autophagy fine tunes epithelial cells' presentation of self-antigens to thymocytes, which leads to the elimination of some populations of self-reactive T cells (
) (Figure 4).
Tissue Homeostasis and Renovation in Health and Disease
Liver
Regardless of the nutritional situation, basal autophagy metabolizes cytoplasmic components to prevent accumulation of degenerated proteins and organelles (Figure 4). Loss of Atg7 in mouse hepatocytes causes marked accumulation of swollen and deformed mitochondria and the appearance of concentric membranous structures consisting of ER. In addition, loss of Atg7 leads to an increased number of peroxisomes and lipid droplets, as well as the formation of protein aggregates positive for p62 and ubiquitin (
). These mutant mice exhibit severe hepatomegaly (i.e., an enlarged liver) and hepatocytic hypertrophy, followed by hepatitis (
). This defective intracellular quality control in the liver appears to be most severe among various types of tissue-specific deletions of autophagy.
What directly causes these abnormalities? Simultaneous deletion of either p62 or Nrf2 dramatically attenuates the liver injury resulting from autophagy deficiency (
,
). These results suggest that persistent activation of Nrf2, due to impaired turnover of p62, can account for most of the pathogenic changes seen in the livers of autophagy-deficient mice.
Autophagy could be a therapeutic target of α1-antitrypsin deficiency. In affected children, misfolded mutant antitrypsin protein accumulates in the liver and causes liver damage and cirrhosis. Both proteasomal and autophagic pathways have been implicated in degradation of the mutant protein. A recent study demonstrated that an autophagy-activating drug, carbamazepine, decreases the amount of the mutant protein and ameliorates hepatic fibrosis in a mouse model of α1-antitrypsin deficiency (
). Such therapeutic strategies could be applied to a wide range of protein conformation disorders, such as Huntington and prion diseases.
Brain
Even under prolonged fasting conditions, the number of autophagosomes does not increase in nerve cells (
) probably because nerve cells are supplied with nutrients from peripheral organs. However, recent studies demonstrated that autophagy can be induced in cortical neurons, Purkinje cells, (
), and hypothalamic neurons during fasting (
). This autophagic response in the hypothalamus induces lipophagy and upregulates agouti-related peptide (AgRP) expression, which increases food intake (
). Accordingly, deleting Atg7 specifically in AgRP neurons reduces body weight and adiposity. However, the opposite finding has been also reported; suppressing Atg7 in the hypothalamus with short-hairpin RNAs causes obesity through the IKK-β/NF-κB pathway (
). Constitutive suppression of autophagy from the embryonic period versus transient suppression in the adult mice may produce different phenotypes, or distinct types of hypothalamic neurons have different autophagy functions.
In contrast to the rather complicated metabolic role of autophagy, the importance of autophagy as a “housekeeper” is easy to envision because neurons are terminally differentiated and do not divide (Figure 4). Mice lacking autophagy in the central nervous system exhibit neurologic deficits, such as abnormal limb-clasping reflexes, locomotor ataxia, and lack of motor coordination. They also display significant losses of large pyramidal neurons in the cerebral cortex and of Purkinje cells in the cerebellar cortex (
,
,
). Polyubiquitinated proteins and p62 accumulate in neurons in almost all regions of autophagy-defective brains, forming inclusion bodies whose size and number increase with aging (
,
,
). Furthermore, mice with autophagy disrupted specifically in Purkinje cells exhibit progressive axonal dystrophy and degeneration of axon terminals, followed by Purkinje cell death and behavioral deficit. These results indicate that autophagy plays a cell-autonomous role in axonal homeostasis (
,
). Unlike the case of autophagy-deficient hepatocytes, the survival of autophagy-deficient neurons is restored only to a small degree by simultaneous deletion of p62, even though protein aggregates completely disappear in neuronal cells, as they do in hepatocytes (
). Thus, neither p62 accumulation nor inclusion body formation explains the neurodegeneration and impaired neurological function of these mutant mice, and it is likely that autophagy plays a more general role in the brain.
Undoubtedly, the housekeeping role of autophagy becomes more evident when neurons are loaded with pathogenic proteins. These proteins include aggregate-prone mutant forms of α-synuclein that underlie Parkison disease and expanded polyglutamine (polyQ)-containing proteins that are responsible for Huntington disease and spinocerebellar ataxia (Figure 4) (
). Indeed, accumulation of autophagic vacuoles has been demonstrated in human neurodegenerative diseases (
,
). To be degraded by the proteasome, substrate proteins must be unfolded prior to delivery into the narrow proteasomal chamber. However, the aggregated proteins are usually resistant to unfolding, and moreover, polyQ fragments could cause malfunction of the proteasome (
). Accordingly, clearance of misfolded, aggregate-prone proteins is highly dependent on autophagy. In fact, pharmacologic upregulation of autophagy has been shown to be effective in reducing neuronal aggregates and slowing the progression of neurological symptoms in animal models, such as Huntington disease and tauopathy models (
).
Heart
Constitutively suppressing autophagy in the heart from the embryonic period does not initially cause an obvious phenotype in mice, but these animals spontaneously develop cardiomyopathy and die after 6 months of age (
). Sarcomere structure is impaired and deformed in mice, and dysfunctional mitochondria accumulate in their cardiomyocytes. This housekeeping role of autophagy is more clearly observed when Atg5 is disrupted specifically in the heart by tamoxifen; sudden ablation of the autophagic function in adult mice immediately causes cardiac hypertrophy and dysfunction, accompanied by the accumulation of ubiquitinated proteins (
). Thus, constitutive basal autophagy is critically important for cardiac homeostasis (Figure 4).
Besides this basal role, autophagy can also be induced in the heart by pressure overload (
,
). Although the animals appear grossly normal at younger ages, constitutive knockout of Atg5 in the heart sensitizes mice to cardiac dysfunction and dilatation following pressure overload (
). This suggests that autophagy induction in the heart could be an adaptive response to hemodynamic stress. Paradoxically, heterozygous deletion of Beclin 1, which partially reduces autophagic activity, improves cardiac function upon pressure overload (
). This suggests that fully activated autophagy could be a maladaptive response, whereas partial, but not complete, suppression of autophagy may be beneficial. Consistent with this hypothesis, partial suppression of autophagy with histone deacetylase (HDAC) inhibitors ameliorates pressure overload-induced cardiac hypertrophy in mice (
).
Skeletal Muscle
Mice with Atg5 (
) or Atg7 (
) disrupted specifically in the skeletal muscle display age-dependent muscle atrophy. Like autophagy-defective cardiomyocytes, their muscle cells exhibit disorganized sarcomeres and the accumulation of p62, ubiquitinated proteins, and deformed mitochondria, confirming the homeostatic role of autophagy in skeletal muscle (Figure 4). Muscle atrophy can be induced by fasting and denervation, but autophagy does not appear to be involved in both of these atrophy processes. Instead, suppression of autophagy exacerbates fasting- and denervation-induced muscle atrophy (
).
Autophagosomes have been frequently recognized as pathognomonic morphological hallmarks of numerous neuromuscular disorders, including Danon disease, X-linked myopathy with excess autophagy (XMEA), and lysosomal storage diseases (Figure 4) (
). Danon disease with an X-linked dominant inheritance pattern is characterized by cardiomyopathy, myopathy, and variable mental retardation. It is caused by mutations in the coding sequence of Lamp-2, which affects lysosomal function or autophagosome-lysosome fusion (
). Patients with XMEA show similar morphological features to those with Danon disease, implying abnormality of lysosomal function (
).
Autophagy is also implicated in the pathogenesis of lysosomal storage diseases. Deleting acid alpha-glucosidase in mice is an animal model of Pompe disease, in which glycogen accumulation in lysosomes causes skeletal myopathy and cardiomyopathy. Autophagy may be one route by which cytosolic glycogen is delivered to the lysosome. Suppression of autophagy in the skeletal muscle of this mouse model reduces the glycogen content, and there is a remarkable additive effect with enzyme-replacement therapy (administration of recombinant human acid alpha-glucosidase) (
). This raises the possibility that suppression of autophagy could be a new therapeutic approach for Pompe disease.
Bethlem myopathy and Ullrich congenital muscular disorder are caused by mutations in any of the three genes coding for the extracellular matrix protein collagen-6. Mice deficient in collagen-6 (col6a1) provide a model for these diseases. Loss of collagen-6 leads to an activation of Akt, which decreases autophagy through inactivation of the transcription factor FoxO3 and activation of mTOR (
). Consequently, structurally altered organelles, including mitochondria and sarcoplasmic reticulum, accumulate in the muscles of the col6a1 knockout mice, causing cell toxicity and death. Of note, induction of autophagy by treatment with a low-protein diet, rapamycin, or cyclosporin A clears the abnormal organelles and ameliorates the dystrophic phenotypes in col6a1 knockout mice (
).
Intestine
Crohn disease is one of the most common inflammatory diseases, and a link to the innate immune system has been established for this disease. However, several studies have identified an unexpected association between Crohn disease and a single-nucleotide polymorphism (SNP) of Atg16L1 (threonine 300 is replaced by alanine) (
,
; The
Wellcome Trust Case Control Consortium, 2007
). To date, this is the only case in which a SNP in the core ATG genes has been associated with a human disease, and the finding highlights a potential importance of autophagy in intestinal biology (Figure 4). Later studies also identified an association between Crohn disease and IRGM (immunity-related GTPase family, M), which has also been implicated in autophagy against intracellular pathogens (
,
).
However, the role of autophagy in the intestine and the pathogenesis of Crohn disease are not simple. Threonine 300 is located in the C-terminal WD repeat domain in Atg16L1. Because yeast Atg16 lacks the entire WD repeat domain, the N-terminal half of Atg16L1 may be sufficient for conventional autophagy, and the role of the WD domain remains unknown. One study suggested that the Atg16L1T300A mutant is defective in autophagic sequestration of intracellular bacteria but not in canonical autophagy (
). Another study, however, suggested that this mutant has intact activity in both canonical autophagy and antibacterial autophagy (
). More experiments are required to determine how the T300A replacement modifies the molecular function of Atg16L1.
Furthermore, evidence from in vivo studies has suggested that autophagy or ATG genes have a role in maintaining the normal function of Paneth cells (
,
). Although complete systemic deletion of Atg16L1 causes neonatal lethality (
), Atg16L1 hypomorphic mice, which express a very low level of Atg16L1, are viable and grossly normal. However, these mice exhibit structurally abnormal Paneth cells with cytoplasmic lysozyme staining. In addition, Paneth cells normally possess granules containing antimicrobial peptides, but in the Atg16L1 hypomorphic mice, these granules are disorganized (
). Crohn disease patients homozygous for the ATG16L1 risk allele (T300A) show similar Paneth cell abnormalities as mice with Atg5 knocked out specifically in the intestine. Furthermore, pathology of the Paneth cell can be dependent on viral infection and probably other environmental factors, as well (
). Thus, autophagy appears to be important for the integrity of vesicle secretion in Paneth cells.
In addition to these cell-intrinsic mechanisms, a link between autophagy and the immune system can also contribute to the pathogenesis of Crohn disease. This link occurs through a direct interaction between Atg16L1 and NOD2/CARD15 (which has the strongest association with Crohn disease) in the bacterial sensing pathway (
) and through the regulation of inflammatory cytokine production from macrophages (
).
Pancreas
Constitutive suppression of autophagic activity in pancreatic β cells causes reduction in β cell mass, hypoinsulinemia, and the accumulation of ubiquitinated proteins, p62, and deformed organelles (i.e., mitochondria and ER) (
,
). Thus, basal autophagy is important for maintenance of β cell volume and function (Figure 4). Furthermore, autophagy can be activated in β cells by free fatty acids. This process is considered to be an adaptive response, particularly in the presence of insulin resistance (
).
By contrast, autophagy may exert a detrimental effect in the exocrine pancreas. Mice with Atg5 deleted specifically in acinar cells grow normally and show no significant abnormalities of pancreatic histology and blood biochemistry, suggesting that basal autophagy may be dispensable in pancreatic acinar cells (
). However, these mice are resistant to acute pancreatitis induced by cerulein (a cholecystokinin analog). Inappropriate activation of trypsinogen inside acinar cells, which is a cause of acute pancreatitis, is reduced in the absence of autophagy (
). Together with the evidence that autophagosomes/autolysosomes accumulate in acute pancreatitis (
,
), these data suggest that trypsinogen could be delivered to the lysosome by autophagy to become the active form in pathogenic settings (Figure 4). However, another study reported an opposite result in which blocking lysosomal function contributed to the pathogenesis of pancreatitis (
).
This apparent discrepancy could be resolved by the findings of future studies. Activity of cathepsin L, which can completely degrade trypsinogen, is severely reduced in pancreatitis. In contrast, activity of cathepsin B, which converts trypsinogen into active trypsin, seems to be relatively spared in the zymogen granule-containing fraction (
). This cathepsin “unbalance” could also explain why acute pancreatitis occurs only under special conditions and not following usual starvation, during which trypsinogen should be completely digested even if delivered to the lysosome.
Kidney
Podocytes (visceral epithelial cells) wrap around the capillaries of the glomerulus in the kidney, which help to filter blood. Podocytes display a high level of basal autophagy, implying that they might rely heavily on constitutive autophagic activity for their homeostasis (
). Consistent with this, podocyte-specific deletion of Atg5 causes glomerulosclerosis with an accumulation of intracellular ubiquitinated proteins in aging mice (
). In addition, these mice show increased susceptibility to proteinuric renal diseases caused by puromycin aminonucleoside and adriamycin, which normally upregulate autophagic activity in intact podocytes (
).
Autophagy also occurs in renal tubules. Mice with Atg5 specifically deleted in the proximal tubules exhibit the accumulation of misfolded proteins and deformed organelles, and they are susceptible to ischemia-reperfusion injury (
). These studies suggest that both podocytes and tubular cells require basal autophagy for homeostasis and adaptive autophagy to cope with stresses (Figure 4).
Lung
Deletion of Atg7 in bronchial epithelial cells leads to the accumulation of p62 and activation of Nrf2. This results in hyper-responsiveness to cholinergic stimuli, suggesting that autophagy has a homeostatic role in lung epithelia (Figure 4) (
). Autophagy also seems to be involved in the development of pulmonary diseases. Cystic fibrosis is a common recessive genetic disease, which is caused by a mutation in cystic fibrosis transmembrane conductance regulator (CFTR), a cAMP-dependent chloride channel. A single amino acid deletion, called ΔF508-CFTR, causes the protein to misfold, leading to premature degradation or aggregation. As a result, secretory organs, such as the lung and pancreas, are affected. Airway epithelial cells from cystic fibrosis patients and mouse models of the disease have reduced autophagic activity likely because Beclin 1 is sequestered into cytoplasmic aggregates (
). Impaired autophagy could be pathogenic because restoration of Beclin 1 expression rescues the cystic fibrosis phenotype in mice.
Bone
Although the physiological roles of autophagy in bone remain largely unknown, autophagy may be related to the development of bone diseases. Paget disease of bone is a chronic and metabolic bone disorder that is characterized by increased bone turnover within discrete lesions throughout the skeleton. Mutations in p62, which is a substrate of selective autophagy, have been frequently identified in Paget disease of bone; these mutations are predominantly in the UBA domain of p62 (
). The p62 mutations cause increased osteoclastogenesis by activating TRAF6–NF-κB signaling (Figure 3) (
). This suggests that the regulation of p62 levels by autophagy is important for bone formation.
Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia (IBMPFD) is a rare inherited disorder that exhibits multiple phenotypic features, including myopathy with rimmed vacuoles and bone disease. This disorder is caused by a mutation in valosin-containing protein (VCP)/p97. Although VCP has multiple functions, such as in ER-associated degradation, it also participates in the maturation of the autophagosome, and immature autophagosomes accumulate in IBMPFD muscles and IBMPFD mutant-expressing mice (
).
Tumor Formation and Progression
A tumor-suppressive role of autophagy has been implicated and appears to be particularly important in the liver. Spontaneous benign tumorigenesis is observed in the livers of mice with systemic mosaic deletion of Atg5 (i.e., the Atg5 gene is deleted in only some populations of cells in various tissues) or with Atg7 specifically disrupted in hepatocytes (
,
). Interestingly, tumors are not formed in other organs in the Atg5 mosaically deleted mice. Enlarged mitochondria, with at least partially impaired functions, accumulate in hepatocytes of Atg5- or Atg7-deficient mice (
,
). As previously demonstrated in other cell types (
Karantza-Wadsworth et al., 2007
,
), the oxidative stress responses and genomic damage responses are enhanced in autophagy-deficient livers. Accumulation of p62, at least partially, contributes to tumor growth because the size of the liver tumors in Atg7−/− mice is reduced by the simultaneous deletion of p62 (
). This may be due to dysregulation of NF-κB signaling (
) and persistent activation of Nrf2 (
). Mice heterozygous for Beclin 1, who have significantly less autophagic activity, are also prone to cancer. These animals develop spontaneous tumors, including hepatocellular carcinoma, and they have an increased incidence of liver tumors after infection with hepatitis B virus (
,
).
Although autophagy functions as a tumor suppressor in nontumor cells or the early stages of tumor cell development, autophagy becomes important for cancer cell survival once tumors are established. Cancer cells have an increased metabolic demand (in terms of both energy source and building blocks) for proliferation, and they often need to grow under hypoxic conditions until angiogenesis is sufficiently established. Therefore, cancer cells, particularly those with Ras mutations, rely heavily on autophagy and are “addictive” to autophagy (
). Growth defects caused by autophagy suppression have also been observed in Myc-induced lymphoma and polyoma middle T-induced mammary tumor cells (
,
). Autophagy can also be an adaptive response to chemotherapy (
). However, the involvement of Ras is not simple; Ras-induced autophagy contributes to tumor suppression by inducing autophagic cell death and senescence (
,
). The Ras-mediated autophagy might have different roles in tumor growth dependent on cellular context or cancer stage.
Given the role of autophagy in tumor progression, autophagy suppression could be a strategy for cancer treatment (
,
). In fact, tumor cell death can be induced in mouse models using drugs that inhibit autophagy (e.g., the lysosome-inhibitory reagent chloroquine) in combination with conventional chemotherapy (
,
,
). After these basic studies were published, >20 clinical trials of autophagy-inhibiting drugs (i.e., hydroxychloroquine and chloroquine) were initiated (http://clinicaltrials.gov/ct2/results?term=autophagy) (
).
One concern could be that, if autophagy is suppressed systemically, it may cause many adverse side effects because autophagy is critically important in almost all tissues. However, thus far, hydroxychloroquine seems to be tolerated (
). It is possible that only a partial reduction in autophagic activity, which may show no apparent effect on normal cells, could be beneficial in human diseases. This may be analogous to the effect of proteasome inhibitors: malignant myeloma cells that secrete immunoglobulin are more dependent on proteasomal degradation than normal cells, and the proteasome inhibitor bortezomib is very effective for the treatment of myeloma (
).
Antiaging: Renovation of the Whole Organism
As autophagy has many effects on cellular renovation, it would be reasonable to assume that autophagy can contribute to whole-body rejuvenation. As discussed above, suppression of autophagy causes age-dependent dysfunction in various organs. Interestingly, many regimes that promote longevity, including calorie restriction, TOR suppression, sirtuin activation, and spermidine treatment, are able to induce autophagy (
,
). The central question is whether this represents simply a correlation or whether autophagy is indeed one of the key effectors of these regimens. Genetic studies performed in C. elegans have shown that some of the autophagy-related genes are required for life-span extension induced by inhibition of insulin/IGF-like signaling and calorie restriction (
,
,
,
,
), although not all autophagy-related genes have a longevity-promoting effect (
). Likewise, autophagy is also required for life-span extension induced by activation of sirtuins (higher eukaryote homologs of the yeast NAD+-dependent deacetylase Sir2) (
), silencing of TOR (
,
), spermidine treatment (
), and p53 suppression (
). These data indicate that autophagy is a common downstream effector in various life-prolonging signaling pathways. However, as other autophagy-independent pathways are also known to be important, how much autophagy contributes overall to the longevity effects of each regimen needs further investigation.
How can autophagy prolong life span? One obvious mechanism is the cell-autonomous function of autophagy, which avoids accumulation of toxic proteins (e.g., misfolded or aggregation-prone proteins) and organelles (e.g., damaged mitochondria). Additionally, autophagy could reduce inflammatory cytokine secretion and spontaneous tumor incidence, which may also account for its longevity-promoting effect (
).
Conclusion
Cells routinely replace their contents to stay healthy but also to make morphological and functional changes. In this Review, we discussed diverse physiological and pathological processes from the perspective of “autophagy” as an intracellular renovation system. Such a multidisciplinary view is useful to understand why this “self-eating” system has been conserved throughout evolution, how it could participate in normal cellular regulation as well as pathogenesis of human diseases, and how we can take advantage of it for disease therapy. However, many fundamental questions remain. Even with the recent development of sophisticated research tools, such as Cre-mediated conditional knockout techniques, the physiological role of autophagy still remains unknown in some key organs, such as in the bone, skin, and blood vessels. Additionally, although selective autophagy substrates have been identified, the physiological significance of degradation of each substrate, particularly of ubiquitinated proteins, needs to be examined further.
Critical issues also remain with regard to autophagy in therapeutics and diagnostics. Effective indicators or biomarkers for autophagy activity are not currently available. Such markers are important to determine autophagic activity in the disease setting, particularly when monitoring drug effectiveness during autophagy-modulating therapy. Furthermore, the autophagy-modulating drugs currently available are not strictly specific, and the development of more specific drugs will be required. Likewise, although the upregulation of autophagy could be theoretically beneficial for eliminating aggregate-prone proteins, damaged mitochondria, and intracellular bacteria, how these selective autophagic pathways can be stimulated is another challenging issue. Nonetheless, the reality of autophagy-modulating therapy is now closer than was ever expected or predicted.
Acknowledgments
We apologize to authors whose work could not be included because of space limitations. N.M. and M.K. are supported by Funding Program for Next Generation World-Leading Researchers.
References
- Al Rawi S.
- Louvet-Vallée S.
- Djeddi A.
- Sachse M.
- Culetto E.
- Hajjar C.
- Boyd L.
- Legouis R.
- Galy V.
Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission.
Science. 2011; (Published online October 27, 2011)https://doi.org/10.1126/science.1211878- Alirezaei M.
- Kemball C.C.
- Flynn C.T.
- Wood M.R.
- Whitton J.L.
- Kiosses W.B.
Short-term fasting induces profound neuronal autophagy.
Autophagy. 2010; 6: 702-710- Amaravadi R.K.
- Lippincott-Schwartz J.
- Yin X.M.
- Weiss W.A.
- Takebe N.
- Timmer W.
- DiPaola R.S.
- Lotze M.T.
- White E.
Principles and current strategies for targeting autophagy for cancer treatment.
Clin. Cancer Res. 2011; 17: 654-666- Amaravadi R.K.
- Yu D.
- Lum J.J.
- Bui T.
- Christophorou M.A.
- Evan G.I.
- Thomas-Tikhonenko A.
- Thompson C.B.
Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma.
J. Clin. Invest. 2007; 117: 326-336- Bence N.F.
- Sampat R.M.
- Kopito R.R.
Impairment of the ubiquitin-proteasome system by protein aggregation.
Science. 2001; 292: 1552-1555- Bjedov I.
- Toivonen J.M.
- Kerr F.
- Slack C.
- Jacobson J.
- Foley A.
- Partridge L.
Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster.
Cell Metab. 2010; 11: 35-46- Cadwell K.
- Liu J.Y.
- Brown S.L.
- Miyoshi H.
- Loh J.
- Lennerz J.K.
- Kishi C.
- Kc W.
- Carrero J.A.
- Hunt S.
- et al.
A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells.
Nature. 2008; 456: 259-263- Cadwell K.
- Patel K.K.
- Maloney N.S.
- Liu T.C.
- Ng A.C.
- Storer C.E.
- Head R.D.
- Xavier R.
- Stappenbeck T.S.
- Virgin H.W.
Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine.
Cell. 2010; 141: 1135-1145- Cao D.J.
- Wang Z.V.
- Battiprolu P.K.
- Jiang N.
- Morales C.R.
- Kong Y.
- Rothermel B.A.
- Gillette T.G.
- Hill J.A.
Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy.
Proc. Natl. Acad. Sci. USA. 2011; 108: 4123-4128- Chan N.C.
- Salazar A.M.
- Pham A.H.
- Sweredoski M.J.
- Kolawa N.J.
- Graham R.L.
- Hess S.
- Chan D.C.
Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy.
Hum. Mol. Genet. 2011; 20: 1726-1737- Chen Y.
- Klionsky D.J.
The regulation of autophagy - unanswered questions.
J. Cell Sci. 2011; 124: 161-170- Degtyarev M.
- De Mazière A.
- Orr C.
- Lin J.
- Lee B.B.
- Tien J.Y.
- Prior W.W.
- van Dijk S.
- Wu H.
- Gray D.C.
- et al.
Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents.
J. Cell Biol. 2008; 183: 101-116- DeNicola G.M.
- Karreth F.A.
- Humpton T.J.
- Gopinathan A.
- Wei C.
- Frese K.
- Mangal D.
- Yu K.H.
- Yeo C.J.
- Calhoun E.S.
- et al.
Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis.
Nature. 2011; 475: 106-109- Ebato C.
- Uchida T.
- Arakawa M.
- Komatsu M.
- Ueno T.
- Komiya K.
- Azuma K.
- Hirose T.
- Tanaka K.
- Kominami E.
- et al.
Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet.
Cell Metab. 2008; 8: 325-332- Eisenberg T.
- Knauer H.
- Schauer A.
- Büttner S.
- Ruckenstuhl C.
- Carmona-Gutierrez D.
- Ring J.
- Schroeder S.
- Magnes C.
- Antonacci L.
- et al.
Induction of autophagy by spermidine promotes longevity.
Nat. Cell Biol. 2009; 11: 1305-1314